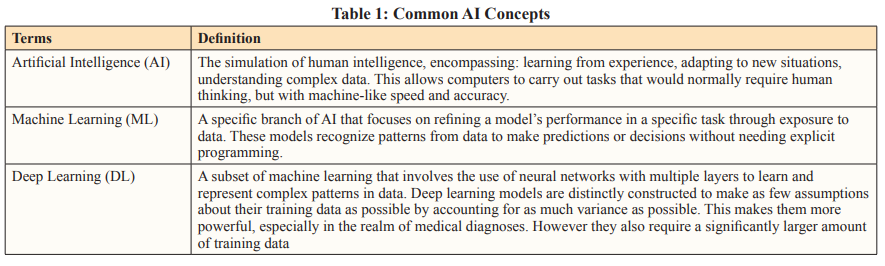
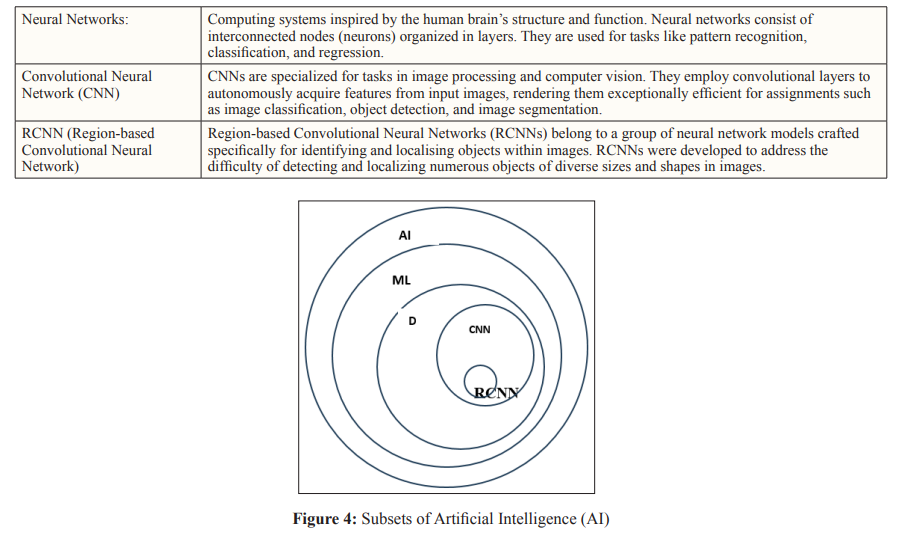
Author(s): Soundarya Kandarpa, Zachariah Chowdhury, Paramita Rudra Pal, Kirti Rajput, and Sahil Ajit Saraf*
In the modern world the number of pathologists are too few to cater to the needs of a growing and ageing population. The complexity of the report and the number of cases per year are constantly increasing. We need innovative solutions to combat this growing gap. In this review paper, we have discussed the basics of digital pathology and machine learning, and have collated a variety of existing AI algorithms, summarizing their applications in the histopathology workflow. We have explored the utility and function of these modules and examined the future trends in the evolving field of artificial intelligence in histopathology
The discipline of pathology is concerned with the origin and nature of disease, underpinning every stage including diagnosis, determining treatment and preventing further affliction [1]. Histopathological analysis in particular, is the gold standard for diagnosing multiple conditions [2]. Often it is only following confirmation from biopsy samples that guideline-adherent management protocols are implemented. Given the indispensable role histopathologists play in the identification of disease, they are required to produce and analyze large amounts of data to arrive at an accurate diagnosis [3]. This data includes glass slide tissue samples with Hematoxylin and Eosin (H&E) staining which are interpreted visually. Although well-established, this method relies on the skill and experience of the pathologist, and is susceptible to disagreement between pathologists due to the subjectivity in interpretation. This could potentially lead to disparities in diagnoses and treatment recommendations [3]. Furthermore, efficiency concerns are compounded by the current shortage of pathologists, their potential fatigue from an increasing workload, and the escalating demand driven by the global surge in cancer cases. These factors collectively contribute to potential delays in the timely review of histopathological samples [4].
The need for more equitable methods of histopathological analyses has led to the emergence of computational technology in laboratories. The digitization of slides by the scanning of conventional glass slides has been adopted since the late 90s. Whole Slide Imaging (WSI) is a digital technology that converts entire glass microscope slides into high-resolution digital images. These images are stored and viewed on computers, allowing pathologists to analyze tissue samples remotely, collaborate easily, and preserve data digitally. These WSIs allow for the acquisition, storage and visualization of high-resolution virtual slides for pathologists to interpret more comfortably and objectively. A high concordance rate between WSI-based frozen section and permanent section diagnosis or on-site interpretation has been demonstrated in a number of studies, further propelling pathology laboratories in their favor. This is especially true in light of the rise of telemedicine and remote pathological analyses [2].
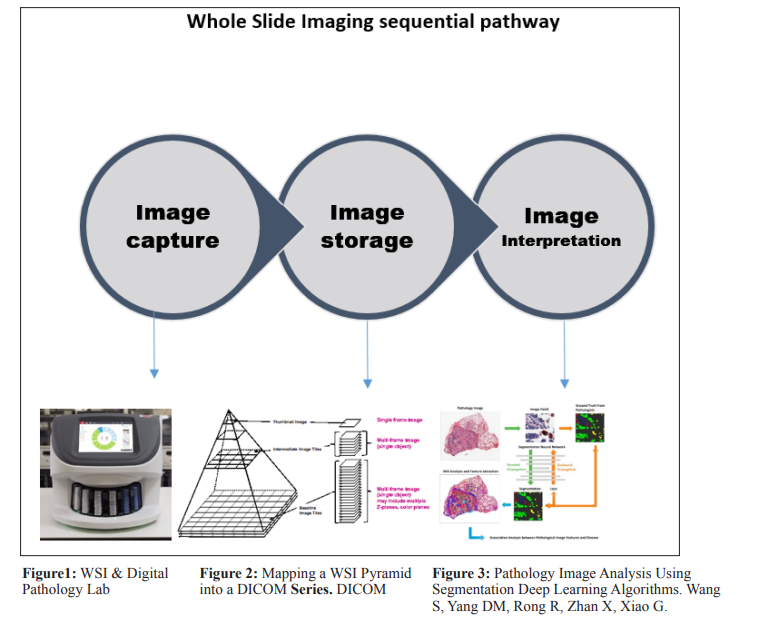
Utilizing WSI technology, there has been promising movement towards the development of Artificial Intelligence (AI) and machine learning algorithms in histopathology for automated or computer-aided diagnosis. Intense research is being undertaken worldwide to develop deep learning software that can successfully recreate the same pattern of image recognition employed by expert pathologists and identify specific diseases at their earliest pathological stage, with a particular focus on malignancies. AI-enabled tools are being developed to predict oncological outcomes from WSIs of H&E-stained tissues, making them a practical alternative to costly genomic testing tools that require sufficient tumor material. AI algorithms have demonstrated the capability to analyze extensive histopathological image datasets with heightened precision. This potential to reduce human errors enhances diagnostic reliability. AI can augment the efficiency of pathologists by prioritizing and pre-screening cases. This targeted assistance enables pathologists to concentrate on intricate cases, potentially elevating overall diagnostic efficiency. AI algorithms remain impartial and unaffected by subjective factors like fatigue or variances in experience, thereby fostering objectivity and standardization in diagnoses.
This review aims to evaluate the latest advancements in the realm of AI in pathology, exploring its potential in providing insights into complex diagnoses and facilitating tailored interventions, thereby improving patient outcomes.
Evaluation of latest studies
Relevant studies were identified from PubMed database initially using the key search terms ‘Artificial intelligence’ and ‘Pathology’. They were then categorized based on the type of research study: (1) Those directly testing their own Deep Learning image analysis models and (2) Meta- analyses evaluating published research of AI in histopathology.
Key Studies: Some of the studies which were evaluated are showcased below in the following tables segregated as per the specific disease
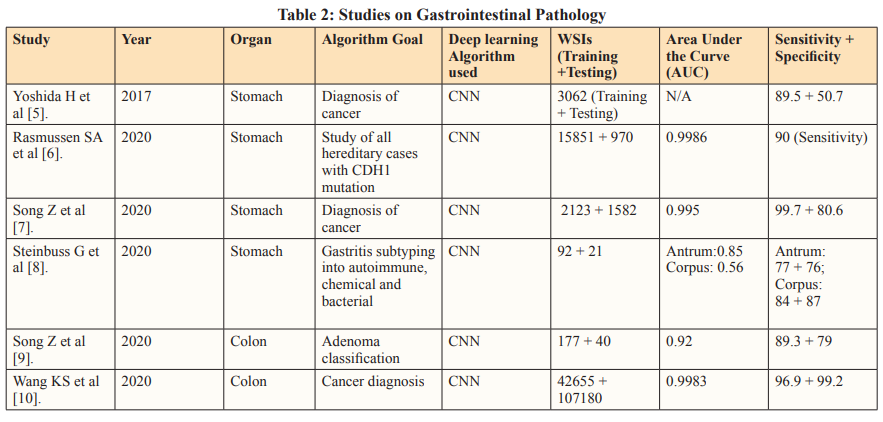
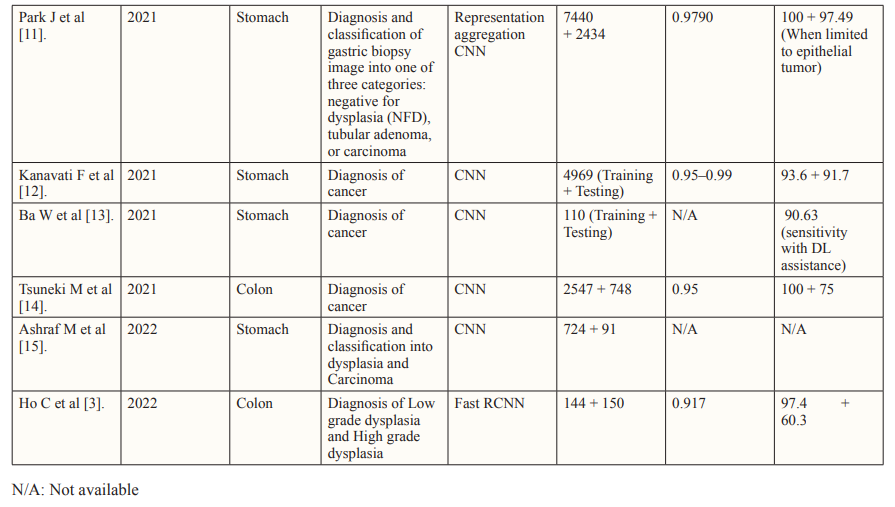
To address overfitting concerns, deep learning models are trained independently using separate datasets. External, geographically distinct testing datasets further validate algorithm performance, enhancing credibility [29]. Innovative Approaches in histopathological analysis such as segmentation model integration, introduce a composite algorithm incorporating glandular segmentation models with machine learning classifiers. This method was utilized by Qritive in the form of a 3-arm- architecture: (i) separating glands from their background, (ii) identifying gland edges, and (iii) instance segmentation, yielding rich and detailed information from the datasets. Segmentation provides detailed information by classifying each pixel, aiding in precise object localization despite computational intensity. Highly skilled pathologists need to make pixel-level annotations on WSIs, a highly time-consuming and expensive task. This is reflected in the relatively small sample size seen in the Qritive study [3].
Heatmaps are a commonly used visual output to show the results of the AI. Although not as informative as segmentation models, the use of Heatmap technology is justified by their streamlined recognition of relevant areas on an image requiring further review [30]. They are hence often used as a part of integrated deep learning models similar to that of the Ibex study (2020) [29,31]. Integrated into deep learning models, heatmaps offer visual insights into predictive model focus areas, aiding in diagnostic endpoint determination. The Ibex study showcases the versatility of its algorithm in not only diagnosing prostate cancer but also differentiating between high-grade and low-grade malignancies. This differentiation, based on the WHO 2016 Gleason Grading system, is achieved with high accuracy, enabling more tailored management strategies and detailed diagnoses. A significant challenge arises during the training stage due to discordance between Gleason grades assigned by pathologists. This discordance leads to misclassifications or the omission of ambiguous and rare cancer variants in datasets. The Gleason score, comprising the sum of the two most prevalent grades in the specimen (primary + secondary), follows distinct criteria for biopsy and prostatectomy. However, variations in interpretation among pathologists contribute to interobserver variability, impacting the consistency and comprehensiveness of AI-assisted diagnoses. Addressing the challenges posed by interobserver variability necessitates refinement of classification systems. Enhancements are crucial for ensuring the accuracy and reliability of AI algorithms in histopathological analysis. The quality of input data plays a pivotal role in training AI models. To mitigate the impact of interobserver variability, efforts should focus on improving the quality and consistency of training datasets, thereby enhancing the robustness of AI-driven diagnostic tools [32].
Recent advancements have significantly enhanced the capabilities of vision classifiers by leveraging Convolutional Neural Networks (CNNs), allowing for a deeper understanding of intricate features and hierarchies within visual data. A recent study focused on Lung subtype Classification utilized a Graph Transformer to augment visual classifiers, demonstrating the effectiveness of incorporating graph-based relationships. By accounting for correlated information, graphs highlight significant connections, facilitating the identification of key predictive features. This approach addresses limitations inherent in patch-based methods,where individual sections of Whole Slide Images (WSIs) are analyzed in isolation, leading to a loss of global context. The fusion of the Graph Transformer and Visual classifier offers a more comprehensive analysis of WSIs, operating at both the patch level and overall-image level [33]. Furthermore, the study introduces a novel class activation mapping technique called Graph-based Class Activation Maps (GraphCAM). Similar to the Graph Transformer, GraphCAM considers the collective influence of various visual elements and their interactions on the network’s final classification. The high congruence between model-identified regions of interest and pathologist-derived assessments underscores the credibility of the model’s outcomes. To mitigate potential noise and variability introduced by patch-level vectors, the study employs the Graph Transformer Processor (GTP) framework. This framework accurately distinguishes between normal WSIs and Lung Adenocarcinoma and Large Cell Carcinoma with high precision. However, the development of the GTP framework, particularly utilizing contrastive learning, is resource-intensive. To optimize efficiency, alternative approaches for defining nodes and generating graphs with improved spatial connectivity should be explored [34].
The segmentation method, tumor probability heatmaps, and patch-level vectors are established deep learning tools that have been used in earlier studies. The success of these tools sparked interest in deep learning technology, as seen in the robustness of recent models. Following the first application of Convolutional Neural Networks (CNNs) in histopathology at ICPR (International Conference on Pattern Recognition) 2012 , numerous studies have explored the potential of deep learning algorithms for analyzing histopathology images, particularly in the context of cancer- related diseases [35-36]. Organizing international challenges has been a highly effective way to promote research in AI for histopathology. These competitions encourage data scientists to compete, showcasing their skills and identifying emerging talents.
CAMELYON16 is one such challenge which stands out as a significant milestone in AI-driven histopathology, focusing on detecting breast cancer metastases in H&E-stained slides of sentinel lymph nodes [37]. This study pioneered a deep learning- based system to detect metastatic cancer in whole slide images of sentinel lymph nodes. To enhance efficiency, the approach excluded background areas using a threshold-based segmentation method, concentrating on cancer-containing regions. The framework consisted of a patch-based classification stage and a heatmap- based post-processing stage. During training, positive and negative patches from whole slide images were utilized to train a supervised classification model. Tumor probability heatmaps were generated using the GoogLeNet CNN architecture, developed by Google in 2014. Post-processing was then applied to compute slide- based and lesion-based probabilities. Remarkably, the algorithm reduced pathologist error by 85%, showcasing the potential of integrating deep learning techniques into the diagnostic workflow. However, the study also highlighted that the AI algorithm’s performance, when used alone, fell short of human experts. This acknowledgment has spurred further research into improving the standalone accuracy of deep learning algorithms, with a particular focus on identifying and mitigating misclassifications.
The CAMELYON16 dataset has been very important for many later studies and challenges, attracting attention from big machine learning companies and even influencing government policies. Public datasets like the Cancer Genome Atlas (TCGA) and the Cancer Image Archive (TCIA) are also available to researchers.
helping them to conduct studies and compare their algorithms against a common standard [38]. Public datasets in the field of deep learning for medical image analysis offer significant advantages but also present certain drawbacks. A key issue is that these datasets may not fully capture the diversity and complexity of real-world data, potentially resulting in biased or overly optimistic assessments of algorithm performance. This limitation underscores the need for diverse and representative datasets to ensure the robustness and generalizability of developed algorithms.
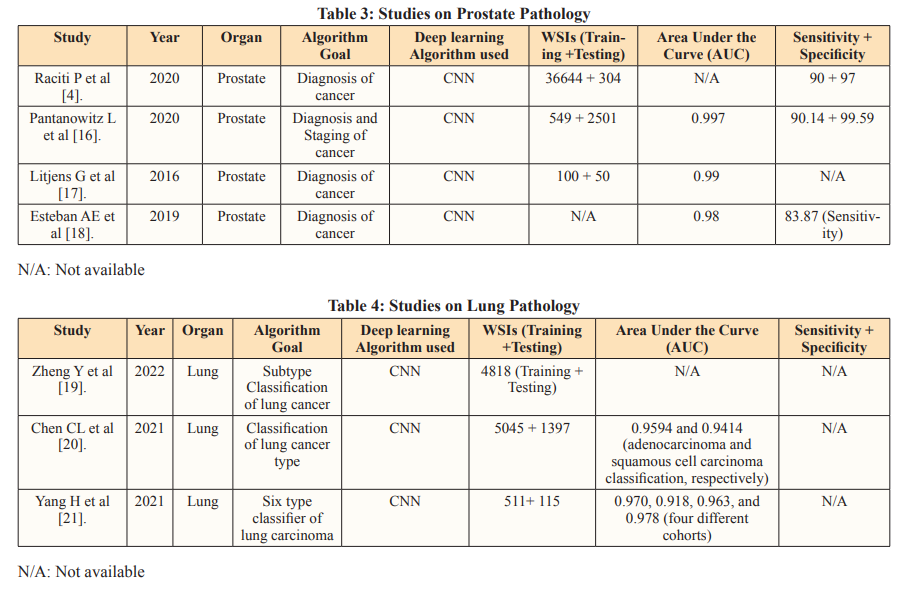
Maintaining uniform data quality and annotations poses challenges with public datasets. The diverse nature of these datasets can lead to inconsistencies in annotation precision and image resolution, potentially influencing the efficacy of both training and evaluation processes. On the contrary, private datasets offer unique insights and control over data collection, but access is typically restricted. These datasets can contain sensitive information about individuals or organizations, making collaboration among researchers or organizations more difficult. It is commendable when studies manage to obtain datasets from multiple unique sources, as it enhances the robustness of their findings.
A China-based study in 2020 went beyond single-site validation by testing a deep learning model on slides collected from two additional hospitals. This approach strengthened the clinical utility of the study. The performance metrics validated the AI model’s reliability and consistent performance across multiple datasets, demonstrating its ability to handle pre-analytical variances created by different laboratories, such as varied sectioning, whole slide imaging (WSI) scanners, and staining configurations. This adaptability to different conditions in real-world clinical settings highlights the AI model’s robustness and potential for widespread clinical application [39].
The latest advancements in genomics have empowered pathologists to discern molecular signatures unique to various types and subtypes of brain cancer. Among these, glioma stands out as the most prevalent and aggressive form, exhibiting distinct subvariants characterized by diverse molecular features influencing their proliferation and metastasis [40-41].
In response to these challenges, a groundbreaking tool named CHARM has been developed, drawing on a dataset comprising 2,334 brain tumor samples collected from 1,524 glioma patients across diverse cohorts. Upon validation with fresh brain samples, CHARM demonstrated an impressive accuracy rate of 93% in pinpointing tumor with specific mutations [41]. Moreover, it effectively classified major glioma types by accounting for their distinctive molecular profiles and responses to treatment. Particularly noteworthy is CHARM’s capability to detect features in the surrounding tissue adjacent to malignant cells, providing insights into the aggressiveness of certain tumor types.
Moreover, the tool unveiled notable molecular alterations in less aggressive gliomas, shedding light on factors influencing their progression, dissemination, and response to treatment. Through correlating cellular morphology with molecular profiles, CHARM attained a level of assessment akin to human interpretation when analyzing tumor samples. This nuanced understanding enhances diagnostic precision as well as the treatment strategies, thereby advancing personalized medicine in the realm of brain cancer management.
Although originally trained on glioma samples, researchers anticipate that CHARM could be customized to address other subtypes of brain cancer. Gliomas, characterized by their intricate molecular profiles and varied cellular appearances, have presented formidable obstacles for AI models compared to more homogeneous cancer types such as colon, lung, and breast cancers [42]. The remarkable performance metrics exhibited by the CHARM tool thus signal a promising frontier for a wide array of pathologies, offering potential advancements in diagnosis, classification, and prognostication across diverse medical contexts.
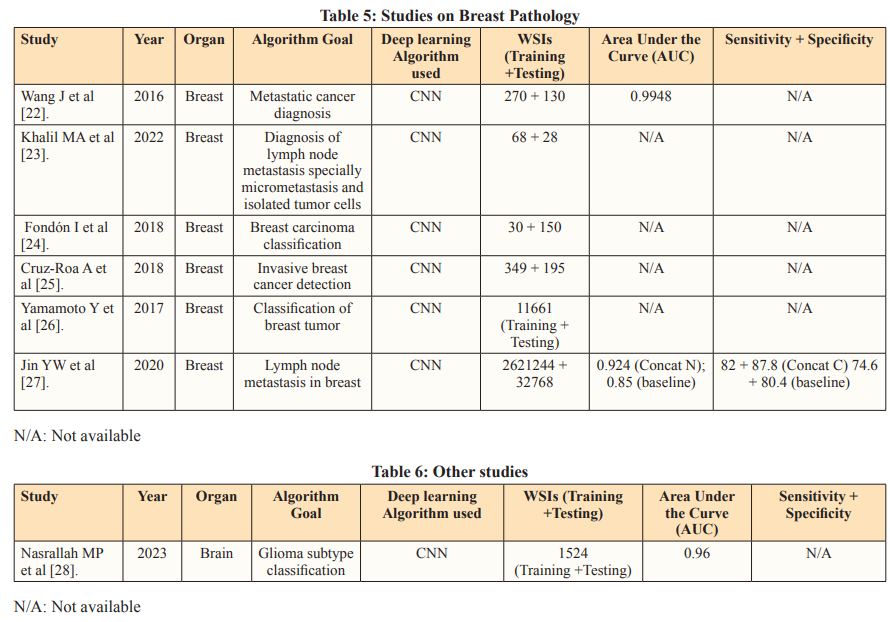
Studies such as Qritive (2022) and Paige Prostate Alpha (2020) prioritize high sensitivity to minimize false negatives, crucial in cancer screening. The AI algorithms aim to assist pathologists by improving diagnostic accuracy without compromising diagnostic specificity [3-4].
Tissue biomarkers play a crucial role in diagnosing, prognosticating, and predicting outcomes for specific subsets of patients. This is especially in the context of the growing shift towards Personalised Medicine. These biomarkers, extracted from tissue samples and primarily interpreted by pathologists, encompass classic factors like histotype, grade, and stage of malignant tumor, as well as newer indicators such as molecular profiles like estrogen and progesterone receptors, and HER2/neu in breast cancer. AI technologies have emerged to bolster the evaluation of tissue biomarkers, particularly in the field of histopathology. AI algorithms analyze complex histopathological data, identifying features that may elude human assessment. This has led to the discovery of AI-based biomarkers capable of predicting treatment responses, somatic mutations, patient survival, and more.

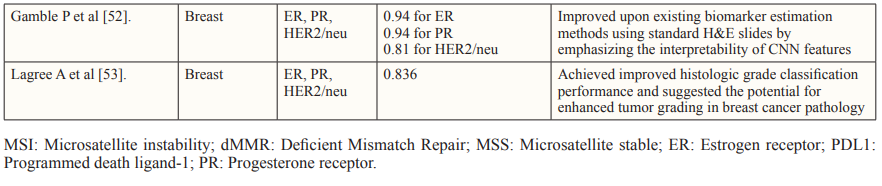
Despite the promising results of the studies discussed, there seems to be a shift from surrogate marker classification such as MSI, to direct prediction of clinical endpoints. This is in order to optimize the production and approval of drugs with hard endpoints in mind and reserve surrogate outcomes for situations of urgency, rarity, or limited treatment alternatives [4].
Additionally, the application of AI in patient selection for immunotherapy, especially in metastatic disease, is a growing area of focus [54]. There is also potential for AI to detect pre-malignant lesions as in Lynch syndrome by being trained to distinguish between somatic and germline etiology, a distinction it presently misses [55]. When compared to next-generation sequencing (NGS), AI can produce significantly more rapid results. However, it cannot currently reliably detect certain mutations and alterations, indicating the need for improvement in classifying other molecular alterations and integrating genomic and histologic data for optimal prediction. In the long term, AI in pathology can lead to cost savings on molecular assays, making it a promising avenue for the future of healthcare.
In the field of pathology, deep learning algorithms are being explored to aid pathologists, but the inner workings of these algorithms are often unknown, making them a “black box” [56]. This means there is no true way to verify the mechanism or pathway the algorithm follows when interpreting data. This raises questions about when AI algorithms can be trusted for critical healthcare decisions, especially in an industry where every clinical diagnosis and decision require justified rationales. A transparent development process is essential for gaining general acceptance of AI technologies within the pathology workflow. Transparency in the data used for testing and training AI models is another consideration. Local training data might not generalize globally, so the transparency regarding data variability can make the pathologists aware about potential performance differences and empower them to adjust their approach accordingly.
Another challenge in histopathology is the inherent inter- observer variability among pathologists. Human experts may interpret and annotate histopathological images differently. AI models, however sophisticated, may not always account for this variability, potentially leading to discrepancies in their diagnostic and prognostic outcomes. This poses a challenge in harmonizing AI-driven analyses with the nuanced expertise of pathologists. Moreover, the issue of new more ambiguous tissue patterns adding to the already limitless variations seen in tissue specimens increases the need to improve the training model for AI frameworks. All Deep Learning architecture is still limited to at least some extent by the annotated data they are trained on, and consistently producing such large annotated datasets proves to be a tedious task for pathologists currently.
AI models trained on one type of tissue or staining technique might not generalize well to other tissue types or stains commonly encountered in histopathology. The field encompasses a wide array of specimens and staining protocols, making it challenging to develop AI models that are universally applicable. Additionally, the computational demands of processing high-resolution histopathological images are substantial, necessitating access to powerful computing resources. This requirement can be a significant limitation for smaller healthcare facilities or regions with limited infrastructure, potentially compromising the scalability and accessibility of AI solutions due to these resource constraints.
Despite these challenges, efforts to improve the generalizability of AI models are ongoing. For instance, training models on diverse datasets that include various tissue types and staining techniques can enhance their robustness. Collaborations between institutions to share data and computational resources could also help address the infrastructure limitations faced by smaller facilities. In this context, the development of more efficient algorithms that require less computational power without compromising performance could further democratize access to advanced AI technologies in histopathology.
As in all industries, but more so in the healthcare industry, data confidentiality forms one of the cornerstone debates in the utilization of AI. One obvious issue with obtaining new datasets is the ethical issues surrounding mining patient data and the crucial question as to whether it is the patients or the pathologists that own the data that is used in these studies. While pathologists would push for the utilitarian use of this data that they painstakingly curate and analyze, the ultimate decision of utilizing confidential data for research and often potentially commercial purposes poses additional hurdles in the way of approval. Despite the rising prevalence of AI in healthcare, patients’ trust in computers to handle crucial decisions including cancer diagnoses remain limited by the lack of accountability if there is an error on the part of the machine. It is essential to have clear accountability hierarchies to ensure patient safety. This accountability extends to AI vendors, pathologists, and healthcare institutions, all of which share responsibility for using AI ethically and effectively in patient care. This conundrum satisfies the Turing test dilemma which suggests that a computer is only as intelligent as its human counterpart when it successfully replaces the human in performing tasks to the extent that it can be considered an imposter [57]. So far all of the AI models discussed have demonstrated value in assistive workflow, where the pathologist is the final evaluator. Currently, the requirement for pathologists to validate and take responsibility for all decisions made by machines undermines their potential as a standalone solution in workplaces experiencing pathologist shortages.
The integration of AI in histopathology demonstrates its prowess as an assistive tool in screening, staging, classification, and prognosis of diseases. The evolution of deep learning technologies offers a multitude of opportunities for model development, allowing developers to prioritize specific features and dictate outcomes accordingly.
Research thus far has shown that segmentation methodologies provide the most detailed insights, although they come with significant computational demands. These models often require skilled pathologists to perform pixel-level annotations, which limits their application due to the associated time and cost constraints. Therefore, it is crucial to recognize the contribution of international challenges that foster competition and innovation to overcome these limitations.
Studies that successfully integrate data from various sources demonstrate the adaptability and utility of AI models across different clinical settings. Biomarker studies have further illustrated that algorithms trained on larger, more diverse datasets tend to have better performance metrics when externally validated. This is particularly important for evaluating the prognostic potential of AI tools in detecting crucial biomarkers.
In essence, the role of AI in histopathology is dynamic and ever- evolving, promising enhanced diagnostic accuracy and valuable support for pathologists. It addresses challenges related to data diversity and model complexity, paving the way for more effective disease diagnosis and management.
