Author(s): Chikodili G Anaukwu*, Blessing I Onyebuagu, Chibuike G Obi, Vivian, N Anakwenze, Chito C Ekwealor, Tobechukwu MC Ajogwu, Victoria I Anyaoha, Josephine C Ohuche, Chidimma Osilo and Amarachukwu B Isiaka
Pseudomonas aeruginosa is notable for its natural bluish-green pigment production, known as pyocyanin. Researchers have credited pyocyanin as an antimicrobial agent, in addition to its other biomedical applications. This study focused on isolating Pseudomonas aeruginosa from the local environment with the ability to synthesize pyocyanin and optimize its cultural conditions for improved yield. A total of thirty-one experimental combinations of process variables using the Central Composite Design (CCD) of Response Surface Methodology (RSM) was ran to find the best conditions for the organism to make pigments. Of the fermentation media evaluated, nutrient broth supplemented with maltose was the best fermentation medium for the organism’s pyocyanin production. A regression model described the interaction between the test variables and the pyocyanin yield with an R2 value of 87.52%, indicating that the model had a satisfactory fitness level. The determined optimal conditions include a maltose concentration of 18 g/L, a pH of 6.8, an inoculum size of 2.3 mL, and an agitation speed of 120 rpm. The optimized condition resulted in a 4.12-fold increase in pyocyanin yield compared to an unoptimized condition. Pyocyanin exhibits high antimicrobial activity compared to the conventional antibiotics assessed against E. coli and Candida albicans. Our study’s findings suggest that statistical models can improve pyocyanin synthesis by Pseudomonas aeruginosa. Furthermore, this pigment has the potential to replace conventional antibiotics in the treatment of microbial infections
Natural pigments are colorants derived from natural sources such as plants, animals, and microorganisms [1]. The demand for biologically safe and environmentally sustainable alternatives drives interest in their production. With the growing global population, there is an escalating need for medications, antimicrobial agents, food, and beverages, as well as a wide range of colored paints, inks, and dyestuffs across multiple industries. Companies often employ synthetic pigments, which frequently pose risks, to meet this demand. Nevertheless, the detrimental consequences of overutilization and improper handling of artificial pigments are rising, presenting significant environmental concerns and grave health issues [2]. Living organisms serve as preferred sources of natural pigments due to their consistent availability, variety, and larger amounts. Mudaliar and Prasad assert that scientists increasingly focus on bacterial pigments such as pyocyanin due to their flexibility, ease of growth, genetic modification potential, and reduced risk of hypersensitive reactions [3].
The genus Pseudomonas synthesizes pyocyanin, a blue-green pigment with antioxidant qualities and a wide-ranging antibacterial effect against bacteria, fungi and parasites [4]. Researchers widely recognize pyocyanin as the most extensively studied member of the phenazines. P. aeruginosa strains exclusively cause the synthesis of the unique, blue-colored pigment. Several studies have utilized pyocyanin as a promising medicinal and biotechnological compound due to its remarkable physicochemical features and biological activities, particularly its antibacterial activity. Pyocyanin damages the cell walls of many microbes and pathogens. This causes harmful oxidative stress-related substances, such as ROS and free radicals, to build up inside the cells [5].
Pyocyanin’s antibacterial action indicates that it has the potential to be a medically useful chemical against various infections. Pyocyanin- based medicines have the potential to enhance the effectiveness of current antibiotics and lower the likelihood of recurring infections. It possesses wound-healing qualities and exhibits anticancer effects [3]. The main way it stops cancer cells from spreading is by making a lot of Reactive Oxygen Species (ROS), such as hydrogen peroxide (H O ) and superoxide (O -). These ROS induce considerable oxidative stress and inflict damage on the cells [6]. Only under specific growth conditions do bacterial species produce bacterial pigments as secondary metabolites [7]. When the cell runs out of resources during the lag phase, it produces pigments. The type of production medium also influences pigmentation [2]. Various bacterial species exhibit distinct growth requirements that result in the production of distinctive colors. To achieve the highest possible amount and quality of pigments, one can simply optimize the media and standardize the incubation conditions [8,9].
Although pyocyanin shows promise in terms of its antibacterial and biological uses, it is crucial to emphasize that further study is necessary to comprehensively comprehend its modes of action, optimize its therapeutic potential, and address any potential safety issues. Also, making pyocyanin-based medicines from microorganisms requires careful consideration of many factors, including fermentation conditions, production dosage, administration methods, and interactions with host cells. In this study, we used response surface methodology to optimize the process conditions for pyocyanin production by Pseudomonas aeruginosa. We then evaluated the pyocyanin’s antimicrobial effects against some pathogenic microorganisms.
We collected greywater samples from multiple restrooms in the students’ dormitories at Nnamdi Azikiwe University, Awka, using sterile sampling bottles. Nnamdi Azikiwe University in Awka has many student dorms, both on and off campus. These hostels provide accommodation for both male and female students. Each hostel structure has restrooms on every floor. The Nnamdi Azikiwe University Awka campus is located on the Enugu-Onitsha Expressway in Awka, Anambra, Nigeria. The university is located in Southeast Nigeria, precisely between latitude 6.245° to 6.283° N and longitude 7.115° to 7.121° E. We accurately labeled the samples with the collection date and promptly sent them to the university’s microbiology laboratory for isolation and analysis.
Isolation and Identification of Pseudomonas Aeruginosa
We used cetrimide agar, a selective medium, to isolate P. aeruginosa. We diluted the greywater sample in a series of 10-fold dilutions and then used the pour-plating procedure to inoculate 1 ml of 10-3 diluted samples onto sterile cetrimide agar plates. We incubated the plates at 25°C for 24 hours to facilitate growth. Following the incubation period, we counted the developed colonies, sub-cultured them on an agar plate, and stored them on nutrient agar slants at a temperature of 4°C. Basic biochemical tests and 16S rDNA sequencing were used for the identification of the recovered isolate. The biochemical tests conducted include Gram’s reaction, catalase test, oxidase test, citrate test, motility test, utilization test, and sugar fermentation test.
The isolate was spread over a clean glass slide, allowed to dry in the air, and then fixed by passing it over the Bunsen burner. After 1 minute, the smear was saturated with crystal violet and then washed with water. Lugol’s iodine was applied and allowed to remain for a further 1 minute. It was then rinsed off and treated with acetone, which functions as a decolorizing agent, for 10 seconds. The slide was treated with safranin dye and then washed after 30 seconds. The slide was air-dried and examined using a × 100 oil immersion lens [10].
A 24-hour-old culture of the test isolate was smeared onto a slide that was devoid of grease, using a sterile inoculating loop. A minimal quantity (2–3 drops) of hydrogen peroxide solution was administered to the smear. The observation of gas bubbles on the slide’s surface indicated a positive outcome [10].
An oxidase test solution was prepared by dissolving 0.1 g of oxidase reagent in 1 ml of sterile water. An aseptic filter paper was submerged in the solution using aseptic forceps and subsequently allowed to air dry. A minute quantity of the 24-hour-old culture of the isolate was deposited onto the filter paper. The presence of a purple color indicated a positive result for the oxidase test [10].
The bacterial isolate was introduced into test tubes with sterile semi-solid nutrient agar by stabbing and then placed in an incubator at room temperature for 24 hours. Observing growth in areas distant from the line of stabbing implied a positive result.
The sugars examined in the fermentation test included lactose, glucose, sucrose, and mannitol. A small amount of the 24-hour-old culture of the test isolate was transferred into a sterile solution containing 1% sugar, 1% peptone water, and bromothymol blue indicator in a test tube with an inverted Durham tube. The tube was placed in an incubator for 24 hours. A change in color from blue to yellow indicated a positive result for sugar fermentation. Additionally, the existence of bubbles at the tip of the Durham tube indicated the formation of gas [10].
The genomic DNA, specifically the 16S ribosomal RNA, was extracted using the ZR bacterial DNA miniprep kit from Zymo Research. The DNA was amplified using the polymerase chain reaction (PCR) with Taq 2X Master Mix from New England Biolabs (M0270). The primers used were the forward primer (27F: AGAGTTTGATCMTGGCTCAG) and the reverse primer (1525R: AAGGAGGTGWTCCARCCGCA). The amplification of the 16S rRNA used a cycling process with the following conditions: an initial denaturation phase at 94 °C for 5 minutes, followed by 36 cycles of denaturation at 94 °C for 30 seconds, annealing at 56 °C for 30 seconds, and elongation at 72 °C for 45 seconds. The final elongation phase, conducted at a temperature of 72 °C, had a duration of 7 minutes. The amplified fragments were sequenced using a Genetic Analyzer 3130xl sequencer from Applied Biosystems, following the manufacturer’s instructions. We used the BigDye Terminator v3.1 cycle sequencing device as the sequencing kit. The genomic analysis was conducted utilizing the Bio-Edit program and MEGA X. The evolutionary history was inferred utilizing the neighbor-joining method, and the evolutionary distances were computed using the Maximum Composite Likelihood methodology [11,12].
The method described in was employed. The initial synthesis of pyocyanin by Pseudomonas aeruginosa was done using four different fermentation media: peptone glycerol broth, nutrition broth supplemented with 1% w/v maltose, nutrient broth supplemented with 1% w/v glucose, and nutrient broth [13]. A 24-hour culture of P. aeruginosa cells (1.2 x 105 cells/ml) was put into a 100-ml Erlenmeyer flask that already had 50 ml of the fermentation media in it. Subsequently, the flask was placed in a rotary shaker set at a speed of 150 revolutions per minute and maintained at a temperature of 25°C.
We separated the supernatant from the fermentation broth after allowing the mixture to develop for 72 hours by spinning it in a centrifuge at a force equivalent to 10,000 times the acceleration due to gravity for 15 minutes and then repeated this process. The supernatant was recovered using the separating funnel and combined with chloroform in a ratio of 2:1. The mixture remained undisturbed for 10 minutes. The solvent layer containing the pigment was extracted using a separating funnel. The solution was acidified with 0.1M HCl and then neutralized with drops of Tris buffer, resulting in the creation of a bluish-green pyocyanin pigment. We collected the generated pyocyanin pigment in a pre-weighed beaker. The pigment was subsequently concentrated through the process of desiccation in a low-temperature oven until it achieved a semi-dehydrated state. The pigment’s weight was determined by subtracting the initial weight of the beaker from its final weight, measured in grams.
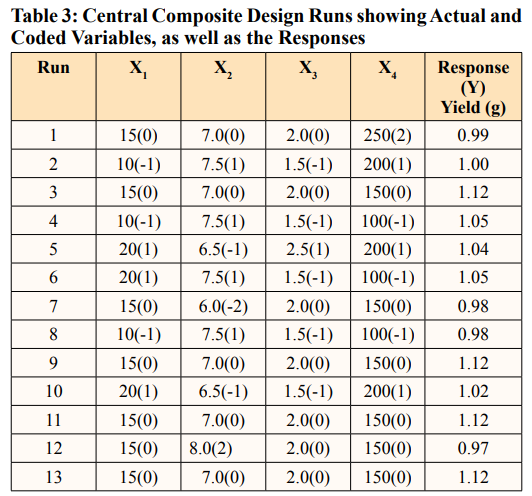
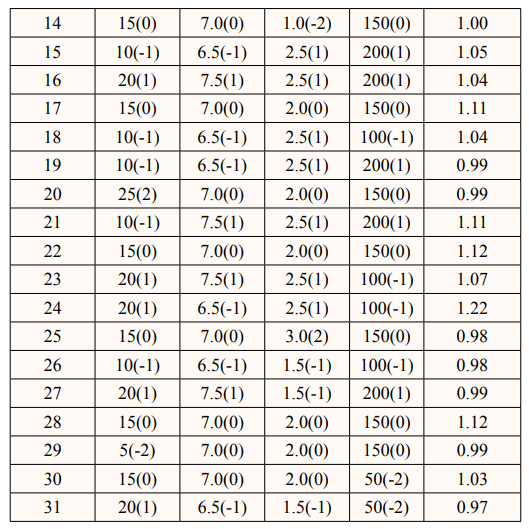
In this study, it was found that the most effective medium for pyocyanin production by Pseudomonas aeruginosa was nutrient broth supplemented with maltose. A Central Composite Design (CCD) of RSM to optimize key media components for enhanced pyocyanin yield was carried out [14]. A 24-full factorial central composite design was employed to establish a second-order polynomial model. The design consisted of four test variables, each with five levels, eight stars, and seven replicates at the center points. The variables evaluated include the maltose concentration (X1) ranging from 5 to 25 g/L, the pH (X2) ranging from 6 to 8, the inoculum size (X3) ranging from 1 to 3 ml, and the agitation (X4) ranging from 50 to 250 rpm. The experiment was devised with Minitab software version 17. We carried out a total of 31 trials, using the weight of the pyocyanin crude extract as the assessed experimental response (Y).
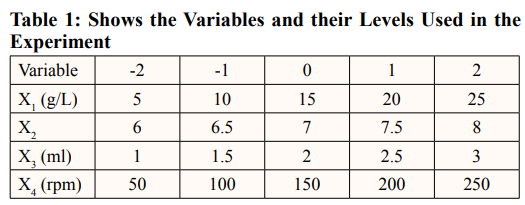
Empirical Validation of the Statistical Model
We further analyzed the obtained results using Analysis of Variance (ANOVA) to determine the influence of each variable on the production of pyocyanin. The R2 value measures the percentage of variance that the considered model can explain. The best conditions were confirmed and recorded as the mean±SD [15]. The response optimizer of Minitab version 17 was used to predict the optimum conditions for pyocyanin production by P. aeruginosa.
The antimicrobial activities of the produced pyocyanin against Escherichia coli and Candida albicans were evaluated using the modified Kirby-Bauer disc diffusion method [16]. We obtained the E. coli and Candida albicans strains for this investigation from the microbiological collection of the microbiology laboratory at Nnamdi Azikiwe University in Awka, Nigeria. We initially standardized the 24-hour-old test organisms using a 0.5 McFarland standard. Subsequently, they were evenly distributed over the sterile Muller-Hinton agar surface. The crude pyocyanin was impregnated onto sterile filter paper (No.1) discs at concentrations ranging from 100 mg/mL to 500 mg/mL. The discs were then placed on the surface of the culture plate containing the test organisms. We then placed the plate in an incubator at 25 °C for 24 hours. The diameter of the zones of inhibition surrounding each pigment disc was measured and recorded in millimeters. The positive controls used in the study were a conventional antibacterial agent, chloramphenicol, and an antifungal agent, fluconazole [10].
The analyses were conducted in triplicate, and the findings were recorded as mean±SD. The mean and standard deviation were calculated using Microsoft Excel 365. The Analysis of Variance (ANOVA) of the responses obtained on CCD was conducted using Minitab software version 20.
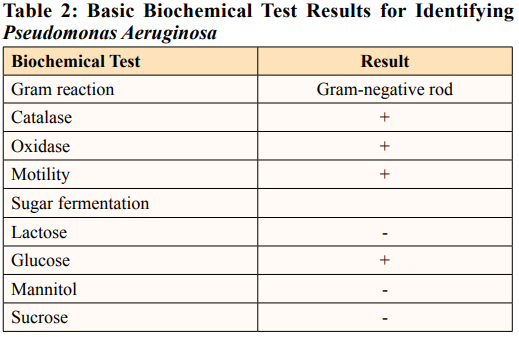
Pseudomonas aeruginosa is the only natural source for pyocyanin pigment production [5]. This study revealed the high potential for pyocyanin production in Pseudomonas aeruginosa, locally sourced from wastewater in Awka, Nigeria. The organism’s preliminary identification revealed a bacterium positive for oxidase, catalase, glucose fermentation, and motility and negative for lactose, sucrose, and mannitol fermentation (Table 2).
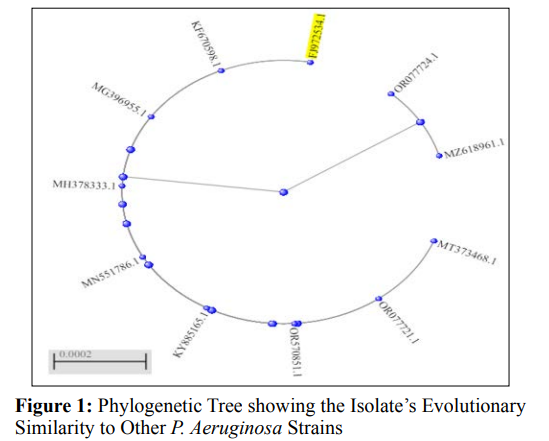
The organism was further identified as Pseudomonas aeruginosa by 16S rRNA sequencing and BLAST analysis, which showed that it was 100% similar to strain NO6 (FJ972534.1). Phylogenetic analysis (Figure 1) presented several P. aeruginosa strains with a pairwise similarity of 100%, such as strain NSJ003.1 (NCBI accession no. MT373468.1), strain GHJ12 (NCBI accession no. MG396955.1), and strain P60 (NCBI accession no. KF670598.1), with 99% similarity to other strains as shown on the phylogenetic tree.
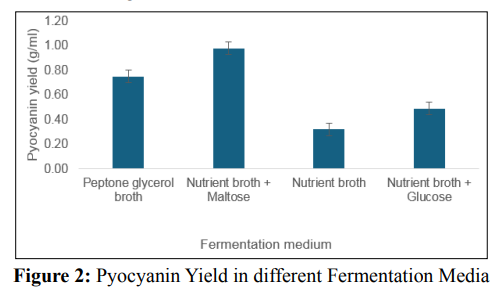
P.aeruginosa can produce pyocyanin in a variety of fermentationmedia, including King’s A medium [5,17]. Luria Bertani medium,nutrient broth, and nutrient broth supplemented with glycerol and/or glucose [17,18]. This study evaluated various media with varying contents, including peptone glycerol, nutrient broth with maltose, nutrient broth with glucose, and nutrient broth, to determine the best medium for pyocyanin production by the organism. Results show that there was a significant difference (P value < 0.05) in the pyocyanin yield in the different media tested. As shown in Figure 2, nutrient broth supplemented with the influence of the four specified parameters on experimental responses [33]. Table 3 shows the central composite experimental design runs that were used to get P. aeruginosa to produce a large quantity of pyocyanin, along with the actual responses that were recorded. Table 3 records thirty-one experimental runs and the pyocyanin yield in grams per milliliter. The resultant regression model is a quadratic polynomial equation that accurately describes the empirical interaction between the test variables and the answer. The equation is as follows: Y = 1.1224 + 0.0108 X1 - 0.0007 X2+ 0.0203 X - 0.0111 X 0.02462 X 2 -0.02837 X 2 - 0.02462 3 4 1 2 maltose gave the highest pyocyanin yield of 0.98 g/mL, followed X 2 - 0.02297 X 2 - 0.0176 X X + 0.0046 X X - 0.0163 X X - 3 4 1 2 1 3 1 4 by peptone glycerol with a 0.75 g/mL yield. The un-supplemented nutrient broth produced the lowest yield. Supplementation of the fermentation medium with adequate carbon and nitrogen sources is a booster for P. aeruginosa’s production of pyocyanin. Research reported sucrose as a much better carbon source than glycerol and glucose. Another study found that mannitol, as a carbon source, yielded the maximum pyocyanin yield, followed by maltose and glycerol [19,20]. It is worth noting that several research groups have attempted to utilize various renewable waste products for pyocyanin production. These include brewing process waste, maize cooking waste, corn steep liquor, potato washing water, coffee waste, tea waste, molasses, cheese waste, grape seeds, taro leaves, pea pods, moss, cotton seed meal, olive waste, vegetable frying oil, corn, soybean, sweet potato, watermelon seeds, and groundnut [17,21-24]. However, this method typically yields relatively low pigment synthesis; therefore, we recommend further research that integrates renewable waste and mathematical models.
Several external variables, such as nutrients, pH, agitation, aeration, and incubation time, influence the synthesis of pyocyanin pigment differently across different strains of P. aeruginosa [25]. Researchers have dedicated their research to genetic engineering methods to enhance pyocyanin yield, aiming to create a more viable strain the culture conditions and medium components can also be optimized to increase an organism’s production yield. Microbes’ susceptibility to fermentation conditions emphasizes the importance of precise process parameters in microbial production [26-30]. To generate pyocyanin, it is critical to carefully examine the process conditions. This research enables the identification of P.aeruginosa’scritical characteristics that influence productivity and optimal pyocyanin yield [31].
Furthermore, it is more precise to use statistical models to determine the interactions and significance of variables to process yield, as well as optimize the process [32]. We utilized Response Surface Methodology (RSM) to develop experimental designs that simultaneously evaluate the interactions between process factors (maltose concentration, pH, inoculum size, and agitation speed) and their impact on production yield. We conducted a multiple regression analysis to build a regression model that examines 0.0105 X2X3 + 0.0077 X2X4 - 0.0056 X3X4, where Y represents the pyocyanin yield measured in grams per milliliter, X1 represents the concentration of maltose, X2 represents the pH level, X3 represents the size of the inoculum, and X4 represents the speed of agitation.
We performed an analysis of variance (ANOVA) to evaluate the statistical significance and appropriateness of the regression model. The findings from Table 4 demonstrates that the regression model exhibited statistical significance (p = 0.05). The lack of significance (P > 0.05) in the lack of fit test shows that the model is good for improved production by the organism. The study showed that the levels of the variables studied did not significantly impact the pyocyanin yield. We evaluated the model’s fit using the regression coefficient (R2) value, which was 87.52%. The calibration of the regression model to the experimental data is appropriate, as it explains 87.52% of the variation between the predicted and observed data. This confirms the model’s suitability for predicting pyocyanin production by P. aeruginosa, which was locally isolated in this investigation under experimental conditions.
The regression model showed that the best conditions for the process were 18 g/L of maltose, a pH of 6.8, an inoculum size of 2.3 ml, and an agitation speed of 120 rpm. These conditions were coded as 0.6263, -0.3838, 0.6263, and -0.5859, respectively. In this study, RSM optimization resulted in a substantial improvement in prodigiosin production output, a 4.12-fold increase compared to the initial fermentation conditions. Previous studies [18,20,34- 36] have recorded the optimization of fermentation media and environmental conditions using response surface methodology to improve the yield of pyocyanin.
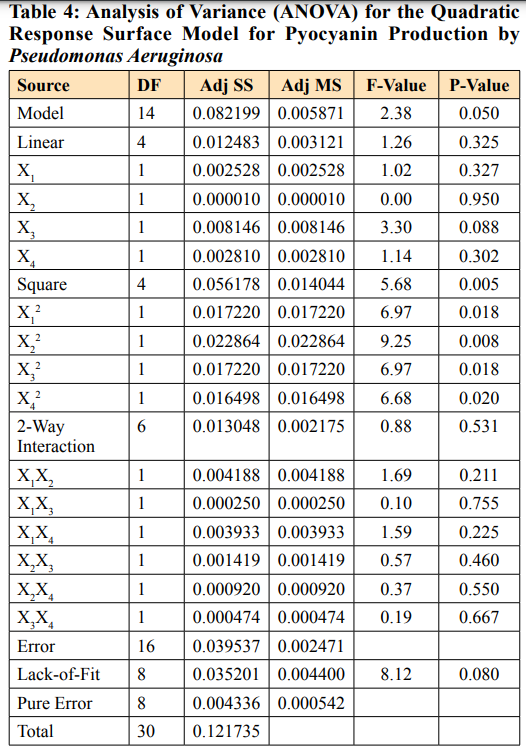
Key: DF stands for degree of freedom, SS for sum of squares, MS for mean square, and P-value ≤ 0.05 is significant at 95 % confidence level. Model Summary: S = 0.0905284, R-sq = 87.52%, R-sq (adj) = 48.21%, R-sq (pred) = 0.00%
The antimicrobial activities of the pyocyanin were assessed using the Kirby-Bauer disc diffusion method. Table 5 shows the results of the antibiotic susceptibility test, which showed the growth-inhibitory zone of different concentrations of pyocyanin pigment against Escherichia coli and Candida albicans. This was compared to chloramphenicol, a common broad-spectrum antibiotic, and fluconazole, a common antifungal. Pyocyanin has shown remarkable efficacy against E. coli, with a zone of inhibition ranging from 1.4±0.01mm to 9.2±0.05 mm, and Candida albicans, with a zone of inhibition ranging from 0.00±0.00 mm to 7.5±0.12 mm. The conventional antibiotics, chloramphenicol, and fluconazole, on the other hand, had antimicrobial effects against the test bacteria and fungi, with inhibition zones measuring 1.2±0.02 mm for chloramphenicol and 2.5±0.01 mm for fluconazole. The antibacterial and antifungal effects of pyocyanin increased with increasing concentration of the pyocyanin (Table 5), and they were much stronger than the effects of the common antibiotics that were tested. However, pyocyanin inhibited bacterial (E. coli) growth more than fungi (Candida albicans) based on the zone of inhibition recorded. The antibiotic effect of pyocyanin recorded in this study corroborates reports in other studies. For example, the pyocyanin produced by P. aeruginosa strains isolated from different clinical specimens was effective against E. coli and Candida species [37]. Several other researchers have recorded a strong antibacterial effect of pyocyanin against E. coli, for example, [21,38, 39]. According to [40], the most sensitive bacteria to pyocyanin is E.coli. They also recorded inhibition of ranges of pyocyanin (250– 300 µg/ml) was recorded and antifungal activity of pyocyanin was better than fluconazole. Another study demonstrated that topical application of pyocyanin in 5 mg/ml concentration eliminate P.albicansin rabbits [41]. On the contrary to our result, high resistance to pyocyanin by E.coli was recorded in a study on the biotechnological and pharmacological assessment of pyocyanin from marine Pseudomonas sp. [42].

In this study, Pseudomonas aeruginosa with the potential for pyocyanin production was isolated from grey water samples in Awka, Nigeria. The response surface method of statistical optimization identified the fermentation condition suitable for production by the isolate and improved pyocyanin yield by 4.12-fold. Pyocyanin produced showed a significantly higher antibacterial and antifungal effect against E. coli and C. albicans respectively than the conventional antibiotics tested. Pyocyanin is thus an excellent alternative to conventional antibiotics. However, the genomic study of the isolate is recommended for enhanced prodigiosin yield.
The Authors wish to acknowledge the Department of Applied Microbiology and Brewing for the provision of laboratory space and resources for the project work.
