Author(s): Young-Gyu Park, Minhyeok Lee, Daeun Kang, Se Jin Park, Wan Jin Hwang, In Beom Jeong, Sun Jung Kwon, Chang Ryul Park* and Ji Woong Son*
BRAF mutations occur in 3-5% of non-small cell lung cancer (NSCLC) patients, with the V600E mutation accounting for about half. Immunotherapy guidelines for NSCLC with BRAF V600E mutations remain unclear due to limited research, in part because of the rarity of the mutation. In this study, we used TCGA data to investigate neoantigens, DNA methylation of mismatch repair (MMR) genes, microsatellite instability (MSI), and immune cell profiling as potential predictors of immunotherapy response. We analyzed data from an entire NSCLC cohort and a subset of adenocarcinomas with prevalent V600E mutations. Neoantigen counts, MMR gene methylation, MSI status, and immune cell profiling were investigated according to the presence or absence of BRAF V600E mutation. The mean age of all patients was 66.2 years. 591 (59.5%) were male, and 513 (51.7%) had adenocarcinoma. Only nine patients (1.0%) had a BRAF V600E mutation. Neoantigen analysis showed no statistically significant differences in insertion/deletion (INDEL) burden; however, a trend toward lower INDEL burden and significantly lower non-synonymous SNP values were observed in the BRAF mutation group. MMR gene methylation, especially MSH2 and MSH6 genes, was significantly higher in the V600E mutation group among entire NSCLC and adenocarcinoma patients. In the non-variant group, four patients showed MSI-H, while all V600E-mutated patients showed MSI-stable (MSS) status. In conclusion, the response to immunotherapy in BRAF V600E-mutated NSCLC is predicted to be limited due to low neoantigen levels (INDEL and SNPs), MSI status, and increased immune cell infiltration associated with low neoantigen load. However, these conclusions rely on retrospective data and require further validation in prospective clinical trials and randomized controlled trials.
Non-small cell lung cancer (NSCLC) exhibits a multitude of oncogenic mutations, prompting extensive research into various treatment modalities, including targeted therapy and immunotherapy. Among these treatments, when the BRAF V600E mutation is identified in NSCLC cases, the National Comprehensive Cancer Network guidelines recommend the use of dabrafenib plus trametinib or vemurafenib/dabrafenib as either first-line or subsequent therapy options [1].
The incidence of BRAF mutations in NSCLC comprises approximately 3-5%, with the V600E mutation constituting roughly half of these cases. Notably, this mutation is predominantly identified in lung adenocarcinoma [2]. Studies conducted in Japan and China have reported even lower frequencies of BRAF mutations, accounting for less than 1%, which is anticipated to be even less common in Asian populations [3, 4].
In cases of advanced or metastatic NSCLC, the primary treatment recommendation involves immunotherapy alone or a combination with cytotoxic chemotherapy, particularly when actionable molecular biomarkers are absent. Additionally, immunotherapy asa standalone approach can be considered for subsequent treatment lines [1]. Regarding EGFR mutations, a prominent oncogenic mutation in NSCLC, it has been established that the combination treatment of EGFR TKIs and immunotherapy does not yield any treatment benefits and is associated with relatively higher toxicity levels [5-8]. However, it’s important to note that there is currently a lack of clear guidelines for immunotherapy treatment in NSCLC patients with the BRAF V600E mutation, and research in this area is also limited.
In the context of BRAF V600E mutated NSCLC, the efficacy of immunotherapy, whether administered in conjunction with targeted therapy (BRAF and MEK inhibitors combination) or as a standalone treatment, remains uncertain. The challenge lies in conducting clinical trials due to the relatively low occurrence of BRAF V600E mutations in NSCLC. Therefore, we aim to leverage the Cancer Genome Atlas (TCGA) data to forecast the response to immunotherapy in NSCLC patients with BRAF V600E mutations [9].
Based on this data, we conducted a comparison between NSCLC patients with BRAF V600E mutations, taking into account their neoantigen counts, DNA methylation of mismatch repair (MMR) genes, and microsatellite instability (MSI) status-all of which serve as markers for predicting the response to immunotherapy. Furthermore, we conducted an analysis of immune cell profiling using Cell-type Identification by Estimating Relative Subset of RNA Transcripts (CIBERSORT).
The TCGA lung cancer dataset comprises a combination of data from the lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC) cohorts. We retrieved baseline clinicopathological characteristics from this database. Gene expression in NSCLC patients was analyzed using the UCSC Xena browser https://xena.ucsc.edu. Subsequently, we obtained BRAF mutation data among NSCLC patients and compared it with the DNA methylation profile of the mismatch repair gene. The DNA methylation data was generated using the Illumina Infinium Human Methylation450 platform and is represented as beta values, ranging from 0 to 1, where higher values indicate increased methylation intensity [10].
Furthermore, we acquired additional data, including MSI status, predicted single nucleotide variant (SNV) and Insertion/deletion (INDEL), neoantigen counts, and CIBERSORT data, from the PanCanAtlas publications’ supplemental data [11,12].
Data analysis was performed by stratifying the entire NSCLC group and the LUAD subgroup, known to primarily express V600E mutations among NSCLCs. We conducted analyses of neoantigen counts, MMR gene methylation values, MSI status, and immune cell profiling based on the presence or absence of BRAF V600E mutations, respectively.
R software version 4.2.0 was used for statistical analysis. The T-test or Wilcoxon-rank sum test was used to compare the two groups for continuous variables. All statistical analyses are judged to be significant when two-sided with p<0.05.
The mean age at diagnosis was 66.2 years, with a range of 33 to 90 years. Among the 993 patients, 591 were male (59.5%), and 513 were diagnosed with adenocarcinoma histology (51.7%). Additionally, 202 patients presented with advanced-stage disease (stage III/IV, 20.3%). In terms of BRAF mutations, nine patients (1.0%) were identified with the V600E mutation, while 49 patients (4.9%) had non-V600E mutations. It’s worth noting that all nine cases of BRAF V600E mutation were associated with adenocarcinoma histology (Table 1).
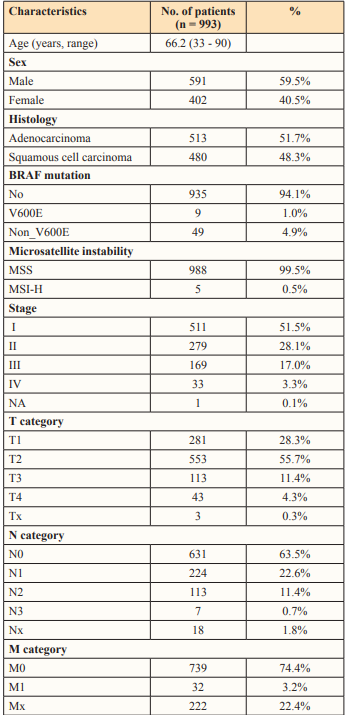
In all NSCLC patients, the median number of INDEL was 8.0 (interquartile range, 4.0-13.0) in the no-variant group and 4.5 (2.0-8.5) in the BRAF V600E mutation group. There was no significant difference between these two groups (P = 0.077). Additionally, the median number of nonsynonymous SNPs was 171.5 (99.0-273.0) in the no-variant group and 47.0 (43.0-90.0) in the BRAF V600E mutation group. Statistically, the number of nonsynonymous SNPs was higher in the no-variant group (P = 0.004) (Figure 1).

Figure 1: Neoantigen counts according to BRAF mutation in all patients. (A) INDEL and (B) nonsynonymous SNP
Within the LUAD cohort, the median number of INDEL was 7.0 (4.0-13.0) in the no-variant group and 4.5 (2.0-8.5) in the BRAF V600E mutation group. No significant difference was observed between these two groups (P = 0.120). Moreover, the median number of nonsynonymous SNPs was 156.0 (65.0-299.0) in the no-variant group and 47.0 (43.0-90.0) in the BRAF V600E mutation group. Statistically, the number of nonsynonymous SNPs was higher in the no-variant group (P = 0.029) (Figure 2).
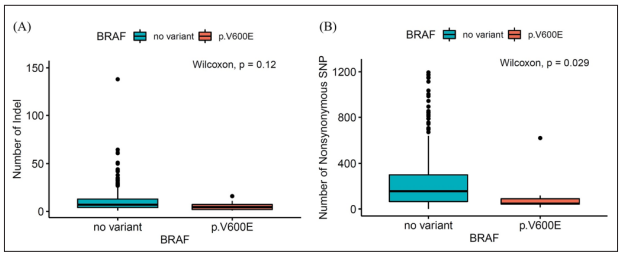
Figure 2: Neoantigen counts according to BRAF mutation in lung adenocarcinoma patients. (A) INDEL and (B) nonsynonymous SNP
In the overall patient group, the median methylation beta values for MSH2 and MSH6 were higher in the V600E group than in the non-variant groups (P < 0.001, P = 0.001, respectively). Similarly, within the LUAD cohort, the median methylation beta values for MSH2 and MSH6 were also higher in the V600E group compared to the non-variant groups (P < 0.001, P = 0.005, respectively). However, both in the entire patient group and in the LUAD cohort, there was no significant difference in methylation values for MLH1 and PMS2 between the V600E mutation group and the non-variant group (Figure 3, 4).
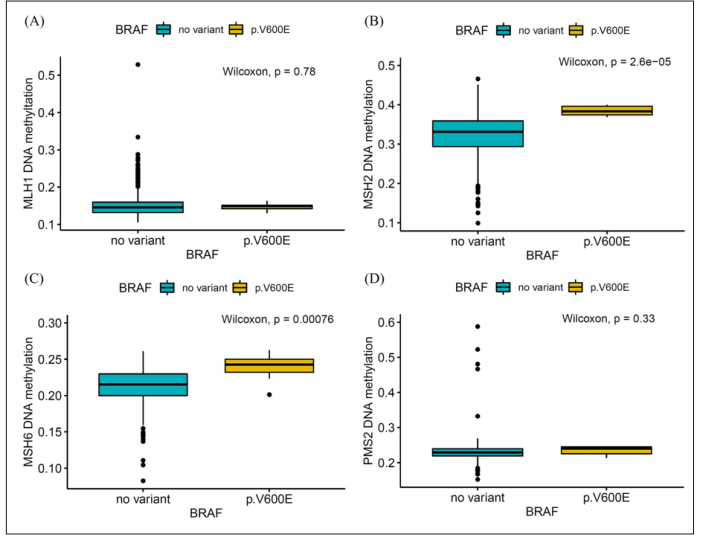
Figure 3: MMR gene DNA methylation values according to BRAF mutation in all patients. (A) MLH1, (B) MSH2, (C) MSH6 and (D) PMS2
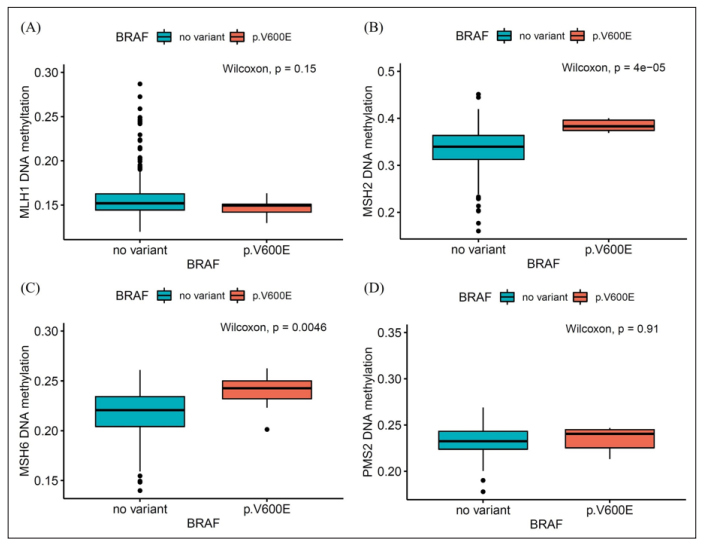
Figure 4: MMR gene DNA methylation values according to BRAF mutation in lung adenocarcinoma patients. (A) MLH1, (B) MSH2, (C) MSH6 and (D) PMS2
Out of a total of 993 TCGA lung cancer patient data, only five patients (0.5%) exhibited a high MSI status (Table 1). Within the novariant group, four patients (0.4%) displayed a high MSI status, while one patient (2.0%) with a high MSI status was identified in the BRAF non-V600E mutation group. Interestingly, in the BRAF V600E mutation group, all nine patients (100%) had an MSI-stable (MSS) status. In the LUAD cohort, only three out of the 513 patients were identified as MSI-H, with two of them belonging to the no-variant group and one to the non-V600E mutant group.
We conducted a comparative analysis of infiltrating immune cells in BRAF V600E mutation tissues and no-variant tissues from NSCLC patients (Table 2). Notably, BRAF V600E mutation tissues exhibited a substantial infiltration of dendritic cells (both resting and activated) as well as resting mast cells when contrasted with the no-variant tissues. Additionally, there was a statistically significant reduction in the infiltration of M0 macrophages in the no-variant group. However, no significant differences were observed between the two groups in terms of CD8 T cells, CD4 memory T cells, regulatory T cells, and memory B cells. These findings were consistent with those obtained from the LUAD cohort (Table 3).
| Immune cell type | CIBERSORT fraction in % of all infiltrating immune cells (mean±SD) | ||
|---|---|---|---|
| No variant (n=926) | BRAF V600E (n=9) | p-value | |
| T cells CD8 | 0.106±0.061 | 0.085±0.050 | 0.303 |
| T cells CD4 naïve | 0.001±0.001 | 0.0±0.0 | 0.585 |
| T cells CD4 memory resting | 0.094±0.069 | 0.109±0.052 | 0.314 |
| T cells CD4 memory activated | 0.009±0.019 | 0.002±0.008 | 0.119 |
| T cells follicular helper | 0.060±0.041 | 0.050±0.043 | 0.385 |
| T cells regulatory (Tregs) | 0.019±0.021 | 0.020±0.020 | 0.678 |
| T cells gamma delta | 0.001±0.004 | 0.0±0.0 | 0.531 |
| B cells naïve | 0.067±0.061 | 0.052±0.059 | 0.356 |
| B cells memory | 0.014±0.030 | 0.025±0.040 | 0.42 |
| Plasma cells | 0.094±0.080 | 0.072±0.090 | 0.213 |
| NK cells resting | 0.013±0.020 | 0.008±0.010 | 0.937 |
| NK cells activated | 0.024±0.027 | 0.036±0.023 | 0.063 |
| Macrophages M0 | 0.079±0.090 | 0.001±0.004 | <0.001 |
| Macrophages M1 | 0.054±0.042 | 0.032±0.040 | 0.068 |
| Macrophages M2 | 0.245±0.102 | 0.262±0.095 | 0.517 |
| Monocytes | 0.023±0.027 | 0.054±0.087 | 0.081 |
| Dendritic cells resting | 0.019±0.033 | 0.046±0.050 | 0.008 |
| Dendritic cells activated | 0.021±0.035 | 0.038±0.032 | 0.022 |
| Mast cells resting | 0.032±0.034 | 0.102±0.054 | <0.001 |
| Mast cells activated | 0.013±0.029 | 0.001±0.003 | 0.091 |
| Eosinophils | 0.001±0.003 | 0.001±0.004 | 0.537 |
| Neutrophils | 0.001±0.018 | 0.002±0.002 | 0.346 |
| Immune cell type | CIBERSORT fraction in % of all infiltrating immune cells (mean±SD) | ||
|---|---|---|---|
| No variant (n=468) | BRAF V600E (n=9) | p-value | |
| T cells CD8 | 0.100±0.058 | 0.085±0.050 | 0.45 |
| T cells CD4 naïve | 0.0±0.0 | 0.0±0.0 | 1 |
| T cells CD4 memory resting | 0.107±0.066 | 0.109±0.052 | 0.77 |
| T cells CD4 memory activated | 0.008±0.020 | 0.002±0.008 | 0.194 |
| T cells follicular helper | 0.058±0.037 | 0.050±0.043 | 0.445 |
| T cells regulatory (Tregs) | 0.023±0.022 | 0.020±0.020 | 0.84 |
| T cells gamma delta | 0.001±0.005 | 0.0±0.0 | 0.479 |
| B cells naïve | 0.067±0.059 | 0.052±0.059 | 0.346 |
| B cells memory | 0.015±0.032 | 0.025±0.040 | 0.502 |
| Plasma cells | 0.093±0.081 | 0.072±0.090 | 0.227 |
| NK cells resting | 0.027±0.016 | 0.008±0.010 | 0.269 |
| NK cells activated | 0.027±0.025 | 0.036±0.023 | 0.182 |
| Macrophages M0 | 0.060±0.074 | 0.001±0.004 | <0.001 |
| Macrophages M1 | 0.047±0.039 | 0.032±0.040 | 0.139 |
| Macrophages M2 | 0.259±0.108 | 0.262±0.095 | 0.831 |
| Monocytes | 0.028±0.032 | 0.054±0.087 | 0.224 |
| Dendritic cells resting | 0.002±0.041 | 0.046±0.050 | 0.026 |
| Dendritic cells activated | 0.019±0.031 | 0.038±0.032 | 0.019 |
| Mast cells resting | 0.040±0.036 | 0.102±0.054 | 0.001 |
| Mast cells activated | 0.007±0.018 | 0.001±0.003 | 0.259 |
| Eosinophils | 0.001±0.003 | 0.001±0.004 | 0.332 |
| Neutrophils | 0.007±0.010 | 0.002±0.002 | 0.3 |
BRAF is a crucial component of the intracellular signaling pathway known as the mitogen-activated protein kinase (MAPK) pathway, which plays a vital role in regulating cell growth and proliferation. When a BRAF mutation occurs, it leads to the continued activation of downstream cell signaling within the MAPK pathway, ultimately resulting in abnormal cell growth and expansion [13,14]. In response to these mutations, targeted therapeutic approaches have been developed. These approaches include BRAF inhibitor monotherapy, aimed at specifically targeting these BRAF mutations, or combination therapy involving BRAF inhibitors and MEK inhibitors, which target the sub-signal transmission system within the MAPK pathway. Historically, patients with NSCLC and the BRAF V600E mutation exhibited a low response rate and poor prognosis when treated with platinumbased chemotherapy. However, the development of such targeted therapies has led to significant improvements in clinical outcomes.
Based on a recently published 5-year updated survival analysis, the combination of a BRAF inhibitor and MEK inhibitor in previously treated or untreated NSCLC yielded promising results. The objective response rate (ORR) was 68.4% (95% confidence interval [CI]; 54.8-80.1) compared to 63.9% (46.2-79.2) in the control group. The median progression-free survival (PFS) was 10.2 months (6.9-16.7) in the combination therapy group and 10.5 months (7.0-14.5) in the control group. Additionally, the median overall survival (OS) was 18.2 months (14.3-28.6) in the combination therapy group and 17.3 months (12.3-40.2) in the control group, demonstrating outstanding survival outcomes [15].
A preclinical study utilizing a BRAF V600E mutant mouse model has provided confirmation that the antitumor activity is enhanced through an immunomodulatory effect when immunotherapy, such as anti-PD-1, is combined with BRAF/MEK inhibitors [16,17]. Building upon this evidence, three randomized clinical trials (KEYNOTE-022, IMspire150, and COMBI-i) were conducted to assess the addition of anti-PD-1 or anti-PD-L1 antibodies to BRAF/MEK inhibitors in patients with unresectable or metametastatic melanoma. These trials demonstrated improved PFS outcomes, with the IMspire 150 trial being the only one to achieve statistically significant PFS results [18-20].
In the case of BRAF V600E mutant NSCLC, there has been a limited number of retrospective studies due to its low prevalence. One such study, a retrospective chart review conducted at Israeli Cancer centers, included 22 patients with BRAF mutant NSCLC who underwent immunotherapy. In this study, the ORR for patients with BRAF V600E mutation was 25%, while it was 33% for those with non-V600E mutations. Additionally, the median PFS for the two groups was 3.7 months and 4.1 months, respectively. Notably, high PD-L1 expression was more prevalent in BRAF mutant NSCLC cases; however, there was no discernible difference in response rates based on PD-L1 expression. Furthermore, the study revealed that BRAF mutant NSCLC patients exhibited low to intermediate tumor burdens, and all of them were classified as having MSS status [21].
Another retrospective study conducted a comparison of immunotherapy outcomes among patients with oncogenic drivers, drawing data from the IMMUNOTARGET registry. This study encompassed 48 patients diagnosed with BRAF mutant NSCLC. The study reported an ORR of 24%, a median PFS of 3.1 months, and a median OS of 13.6 months. Notably, PFS benefits were more pronounced in smokers, and it’s worth mentioning that
the BRAF V600E group exhibited a numerically lower median PFS of 1.8 months in contrast to the 4.1 months observed in the non-V600E mutant group. However, it is important to exercise caution when drawing conclusions from these findings due to the limited sample size [22].
In colorectal cancer, the MSI-H phenotype, without a germline mutation of the MMR gene, is primarily attributed to the silencing of the MLH1 gene via CpG island methylation of its promoter. Importantly, it has been extensively documented that MLH1 DNA methylation is notably high in cases featuring the BRAF V600E mutation [23-25]. This observation aligns with the broader understanding that immunotherapy tends to yield favorable responses in cases characterized by MMR deficiency or MSI-H tumors.
Neoantigens, which are tumor-specific antigens originating from tumor mutations, play a critical role in enabling T cells to detect cancer cells and trigger an anti-cancer immune response [26,27]. These neoantigens can arise from various potential sources, including SNVs leading to nonsynonymous substitutions, INDEL, spliced peptides, translocations, and posttranslational modifications [28]. Notably, prior research has highlighted that tumors exhibiting a high burden of nonsynonymous mutations and frameshift INDEL tend to exhibit more favorable responses to immunotherapy [29,30].
In this study, we observed higher MMR gene methylation values, specifically MSH2 and MSH6, among patients with the BRAF V600E mutation. However, there was no discernible difference in the overall MMR gene methylation values. Additionally, in our neoantigen analysis, we did not find any statistically significant differences in the INDEL burden. Nevertheless, there was a numerical trend toward lower INDEL burden in the BRAF mutation group, and the nonsynonymous SNP value was significantly lower in this group. It’s noteworthy that all patients in the BRAF mutant groups were identified as having MSS status. In the TCGA dataset, a higher neoantigen load was found to be correlated with an increased presence of CD8 T cells, M1 macrophages, and CD4 memory T cells, while there was a lower abundance of Treg cells, mast cells, dendritic cells, and memory B cells across various tumor types [31]. However, our analysis did not reveal any significant differences in the levels of CD8 T cells, M1 macrophages, CD4 memory T cells, Treg cells, and memory B cells between the groups with or without the BRAF V600E mutation. Interestingly, we did observe a notable increase in the populations of dendritic and mast cells within the V600E mutation groups.
In conclusion, the predicted efficacy of immunotherapy application in BRAF V600E mutant NSCLC is limited due to the low level of neoantigen burdens, such as INDEL and SPN, MSI status, and higher immune cell infiltration that are associated with the low neoantigen load. Consequently, the current standard treatment involving BRAF/MEK inhibitors is the recommended approach. However, this conclusion is limited to retrospective data. Prospective clinical trials and RCTs are necessary to further evaluate this recommendation.
Not applicable.
This work was supported by Konyang University Myunggok Research Fund of 2020
Availability of Data and Materials Some data shown here is generated from the original data from the TCGA Research Network https://www.cancer.gov/tcga.
All authors (YP, ML, DK, SJP, WJH, IBJ, SJK, CRP and JWS) designed the study. ML and YP analyzed the data. IBJ and SJK contributed to the interpretation of the data. JWS contributed to the acquisition. ML and YP drafted the manuscript. DK, SJP and WJH revised the manuscript, and CRP and JWS mainly reviewed the manuscript. All authors have read and approved the final manuscript
Not applicable
Not applicable
The authors declare that they have no competing interests.
