Author(s): Suyash Bhogawar*, Deepak Singh, Dwith Chenna and Manasi R Weginwar
Reproducibility is a key component of scientific research, and its significance has been increasingly recognized in the field of Neuroscience. This paper explores the origin, need, and benefits of reproducibility in Neuroscience research, as well as the current landscape surrounding this practice, and further adds how boundaries of current reproducibility should be expanded to computing infrastructure. The reproducibility movement stems from concerns about the credibility and reliability of scientific findings in various disciplines, including Neuroscience. The need for reproducibility arises from the importance of building a robust knowledge base and ensuring the reliability of research findings. Reproducible studies enable independent verification, reduce the dissemination of false or misleading results, and foster trust and integrity within the scientific community. Collaborative efforts and knowledge sharing are facilitated, leading to accelerated scientific progress and the translation of research into practical applications. On the data front, we have platforms such as openneuro for open data sharing, on the analysis front we have containerized processing pipelines published in public repos which are reusable. There are also platforms such as openneuro, NeuroCAAS, brainlife etc which caters to the need for a computing platform. However, along with benefits these platforms have limitations as only set types of processing pipelines can be run on the data. Also, in the world of data integrity and governance, it may not be far in the future that some countries may require to process the data within the boundaries limiting the usage of the platform. To introduce customized, scalable neuroscience research, alongside open data, containerized analysis open to all, we need a way to deploy cloud infrastructure required for the analysis with templates. These templates are a blueprint of infrastructure required for reproducible research/analysis in a form of code. This will empower anyone to deploy computational infrastructure on cloud and use data processing pipeline on their own infrastructure of their choice and magnitude. Just as docker files are created for any analysis software developed, an IAC template accompanied with any published analysis pipeline, will enable users to deploy infrastructure on cloud required to carry out analysis on their data.
In psychology, neuroscience and related sciences, initiatives to improve study reproducibility have reemerged in recent years. The cornerstone of a strong basic research base - one that will support novel hypotheses based on reliable findings and successful technological innovation-is reproducibility. The challenges to reproducibility have become more obvious as a result of the increased emphasis on it, and new tools and techniques have been developed to get around them.
Reproducible Neuroscience research refers to the practice of conducting scientific studies in the field of Neuroscience in a way that allows other researchers to independently verify and replicate the findings [1]. This approach emphasizes the transparency of research methods, data sharing, and the availability of detailed protocols and analyses. The concept of reproducibility has gained significant attention in recent years due to concerns about the credibility and reliability of scientific research across various disciplines, including Neuroscience.
The origin of the reproducibility movement can be traced back to the growing recognition of the “reproducibility crisis” in science. Several high-profile studies in various fields, including Neuroscience, have failed to be replicated, raising questions about the validity and robustness of the original findings. This crisis has prompted researchers and scientific communities to reevaluate their practices and strive for more rigorous and transparent research methods.
The need for reproducibility in Neuroscience research arises from the importance of building a solid foundation of knowledge upon which further research can be based. Reproducible studies allow for the independent verification of findings, reducing the likelihood of false or misleading results being perpetuated [2]. By ensuring that research findings are reliable and replicable, the field can advance with greater confidence, leading to more accurate conclusions and the development of effective treatments and interventions.
However, the adoption of open techniques for transparent, repeatable, and collaborative science is still in its infancy, according to empirical studies of the way laboratories carry out their research [3]. The overwhelming evidence supporting the importance of these procedures and their advantages for particular researchers, the advancement of science, and society at large contradicts this. The knowledge needed to apply open scientific techniques during the various phases of a research project is now dispersed among numerous sources. Making sustainable decisions while navigating the ecosystem of tools can be challenging for even seasoned experts in the field.
Neuroimaging experiments result in complicated data that can be arranged in many different ways. Historically, data were organized differently between institutions and even within a lab. This lack of consensus (or a standard) could lead to misunderstandings and suboptimal usage of various resources: human (e.g., time wasted on rearranging data or rewriting scripts expecting certain structure), infrastructure (e.g., data storage space, duplicates), and financial (e.g., disorganized data has limited longevity and value after first publication). Finally, and most importantly, it produces poor reproducibility of results, even within the lab where data were collected. Therefore, the need for a data standard in the neuroimaging community became essential.
One should mention a community-led standard for classifying, characterizing, and exchanging neuroimaging data, called the Brain Imaging Data Structure (BIDS). MRI, MEG, EEG, intracranial EEG, PET, microscopy, and imaging genetics are just a few of the neuroimaging modalities supported by the constantly developing BIDS standard [4-7]. Numerous more extensions are actively being developed, including fNIRS, motion capture, and animal neurophysiology. The BIDS specification outlines how to arrange the data, which is typically based on straightforward folder structures and file formats (such as NIfTI for tomographic data and JSON for metadata). Through community-driven methods, this specification can be expanded to include new neuroimaging modalities or sets of data types [8-10].
To make it simple for researchers to integrate BIDS into their existing workflows, maximize reproducibility, enable efficient data sharing, and support good data management practices, numerous applications and tools have been provided. For instance, BIDS converters (such as MNE-BIDS for MEG and EEG, dcm2bids, ReproNim’s HeuDiConv and ReproIn for MRI, and PET2BIDS for PET) make it simpler to convert data into BIDS format. Researchers can use the BIDS validator to check that their converted dataset is BIDS-valid.
Users require tools that make it easy to engage with the data once it is in BIDS. The software programs PyBIDS and BIDS-Matlab are two that are frequently used [11]. These tools make it easier to programmatically get particular files, such as all functional runs for a particular subject, as well as relevant dataset queries like how many participants are included in a dataset or what tasks were completed. Finally, BIDS apps are containerized analysis pipelines that output derivative data using entire BIDS datasets as their input [12]. MRIQC, fMRIPrep, and PyMVPA are a few BIDS applications that can be used for MRI quality control, fMRI preprocessing, and statistical learning studies of big datasets, respectively [13].
Evidently, making data available to the community is crucial for reproducibility, enables researchers to learn and teach others to reuse data, develop new analysis techniques, advance scientific hypotheses, and combine data in mega- or meta-analyses. It also allows for more scientific knowledge to be obtained from the same number of participants.
Meanwhile, better explanations of model assumptions, restrictions, and validation are also necessary to increase the impact of modeling across the neuroscience disciplines. Modelers must thoroughly document the assumptions and the methodology for model development and validation when theoretical or conceptual restrictions are driving the development of the model in order to increase transparency and rigor. Better reporting is required for data-driven models in terms of the data that were used to constrain model development, the specifics of the data fitting procedure, and whether or not findings are robust to minute changes in conditions. Better methods for parameter optimization and the investigation of parameter sensitivity are required in both situations.
It is well acknowledged by the research community that, as it was mentioned above, “poor reproducibility of research results is a serious challenge - known as “the reproducibility crisis” - that hinders growths of knowledge and innovation on the one hand and leads to inefficient use of resources on the other hand” [14]. For doing extensive data analysis, an increasing number of biomedical researchers and organizations rely on cloud-based technology. While providers of cloud computing infrastructure exist, there is a significant gap in the availability of scalable cloud tools and technology designed primarily for researchers and deployable in academic institutions. As more and more open tools and infrastructure become essential to researchers, a need to develop sustainability models and resilient strategies to ensure their long-term availability arises.
Hazan et al. provide BindsNET, a Python package for quickly building and simulating such networks for implementation on various CPU and GPU platforms, promoting reproducibility across platforms, for spiking neuron networks specifically geared toward machine learning and reinforcement learning tasks [15].
Blundell et al. concentrate on a method that uses high-level descriptions of complicated models to solve the issues with repeatability that develop as model complexity rises. For simulation and visualization, these high-level specifications must be translated into code, and using code generation to do so automatically improves standards [16]. The authors present a summary of the current code production pipelines for the most popular simulation platforms, multiscale model description languages independent of simulators, neuromorphic simulation platforms, and collaborative model development communities.
Abe et al. rightly claim that the creation of robust, all-purpose data analysis that processes huge datasets is a crucial component of neuroscience research [17]. Unfortunately, consumers of analysis are discouraged from using new data analyses because of a concealed dependence on complicated computing infrastructure (such as software and hardware). Even while open-source software for existing studies is being distributed more frequently, the infrastructure and expertise required to deploy these analyses effectively continue to be major usage hurdles.
Meanwhile, as Saunders rightly claims, “the most pressing problems in science are neither empirical nor theoretical, but infrastructural” [18]. Decentralized digital infrastructure is the best means of alleviating the harms of infrastructural deficits and building a digital landscape that supports, rather than extracts from science.
The immediate effects of neglected infrastructure are well known to the neuroscience community: for each novel analysis, users must invest time and money in hardware setup, software troubleshooting, unexpected breaks during extended analysis runs, processing limitations due to constrained “on-premises” computational resources, and more (see Figure. 1). However, neglected infrastructure has important and urgent scientific ramifications that go far beyond simply being an inconvenience. Reproducibility of infrastructure impact analyses stands out the most. It is quite challenging for analysis developers to work reproducibly when data analytics become more dependent on sophisticated infrastructure stacks [19].
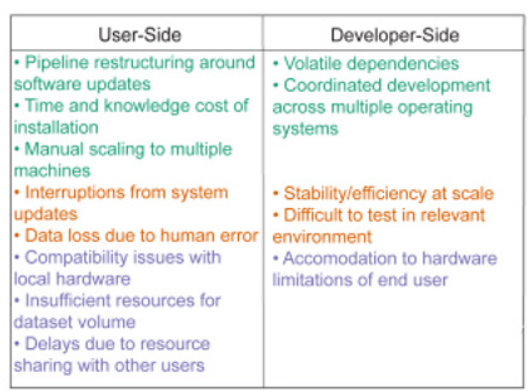
Figure 1: Common problems in neuroscience data analysis [20].
Reproducibility in Neuroscience research offers several benefits to the scientific community and society as a whole. Firstly, it fosters a culture of trust and integrity, enhancing the credibility of scientific findings. Researchers can have greater confidence in building upon previous studies, knowing that the results are reliable. Additionally, reproducibility promotes collaboration and knowledge sharing within the scientific community, as researchers can work together to replicate and validate findings. This collaborative approach facilitates scientific progress and accelerates the translation of research into real-world applications.
Turning to the current landscape of reproducibility in Neuroscience, researchers now have access to a range of tools and resources that support reproducible practices. These tools include data management platforms, version control systems, and open- source software for analysis and visualization. Such tools enable researchers to document and share their research workflows, making it easier for others to replicate the study [21, 22]. Furthermore, initiatives like the Neuroscience Information Framework (NIF) and the International Neuroinformatics Coordinating Facility (INCF) provide platforms and methodologies for data sharing, standardization, and collaboration in the field of Neuroscience. These platforms promote the adoption of reproducible practices by providing a centralized hub for researchers to access and contribute to shared datasets and methodologies.
Given the variety of processing pipelines in neuroscience and requirements to run within geographic and other boundaries, it is imperative that we should enable researchers to reproduce the processing pipelines with their choice of computing infrastructure. However, managing and standardizing various patterns and practices throughout the world will not be practical. To go around the problem we propose a way to standardize computing infrastructure deployment for neuroinformatics on public clouds such as AWS, Azure and GCP. There are standardized cloud services which can act as building blocks for storage, compute, IDEs, custom hardware availability etc.
In this context, special attention should be paid to IaC. According to experts, “the value of IaC is based on 3 pillars: price, speed, and risk reduction. The reduction in costs refers not only to the financial component, but also to the amount of time spent on routine operations. The principles of IaC allow not to focus on the routine, but to deal with more important tasks” [23]. IaC provides stable environments quickly and in a well-organized manner. Development teams do not need to resort to manual configuration- they ensure correctness by describing the required state of the environments using code. Infrastructure deployments with IaC are repeatable and prevent runtime issues caused by configuration drift or missing dependencies. IaC completely standardizes the infrastructure setup, which reduces the possibility of errors or deviations.
Modern IaC is becoming more complex and intelligent. IaC tools will evolve in the direction of expanding functionality and capabilities. They will include increasingly more intelligent features such as automatic error detection and infrastructure optimization [24]. IaC is already being used to manage virtual machine infrastructure, but is already expanding into new areas such as network infrastructure management, container and microservice management, event infrastructure management, and others.
First of all, it is expedient to explore the benefits of adopting Cloud Infrastructure as Code (IaC) in the field of Neuroinformatics research. By enabling end users at the pipeline development stage, leveraging specialized hardware and frameworks, and harnessing the advantages offered by cloud services, researchers can enhance their neuroscientific investigations [25]. The following points outline the benefits of Cloud IaC in this context:
a. Easy Deployment and Built-in Guard Rails: Infrastructure provisioning, software deployment, and configuration management can be automated through IaC tools. Researchers can define their infrastructure requirements as code, ensuring consistent and reproducible deployment across various stages of the research pipeline. Additionally, built-in guard rails provided by cloud platforms enable researchers to enforce security, compliance, and best practices within their infrastructure.
b. On-the-Go User Management: Collaborative neuroinformatics research often involves multiple team members or external collaborators. Cloud IaC allows researchers to easily add or remove users, assign permissions, and manage access control, facilitating seamless collaboration and resource sharing among research teams.
c. Data Governance and Security: Cloud platforms provide robust data governance capabilities, including access controls, data encryption, and compliance frameworks. By leveraging these features, researchers can ensure the security, privacy, and integrity of their data assets throughout the research lifecycle. Furthermore, cloud storage options enable efficient data management, backup, and archiving, alleviating the burden of local storage constraints.
d. F.A.I.R[32]: Every element comprising the Infrastructure as Code (IAC), encompassing storage, computation, networking, and related components, exhibits the capacity for lineage tracking and reproducibility. Given that these fundamental notions align with the FAIR principles, the utilization of IAC can effectively facilitate the FAIR-compliant advancement of informatics pipelines.
e. Moreover, availability of Specialized Frameworks should be especially mentioned. Neuroinformatics research often requires the utilization of specialized frameworks and software libraries for tasks such as neuroimaging analysis or machine learning. Cloud IaC enables researchers to leverage pre-configured environments (docker, singularity images) that include these specialized frameworks, ensuring compatibility and efficient utilization of domain-specific tools. This accessibility allows researchers to focus on the scientific aspects of their work rather than investing significant time and effort into environment setup and dependency management. The overall concept is given on Figure 2.
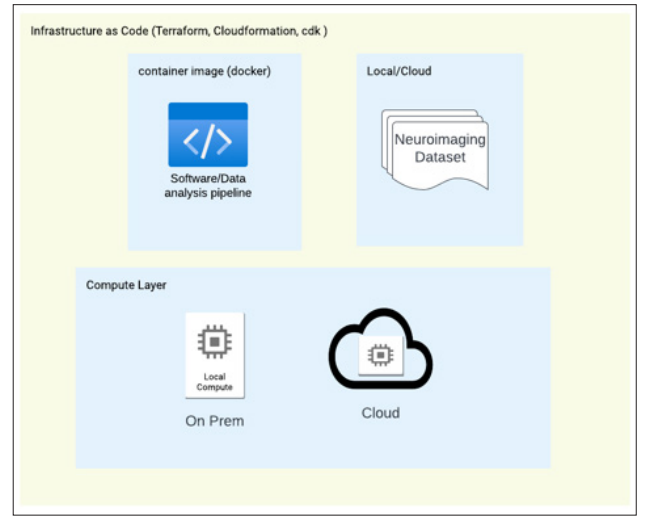
Figure 2: The Concept of Infrastructure as Code.
In particular, in NeuroCAAS users can choose from a list of supported methods (which is continuously updated at www.neurocaas.org), choose a corresponding configuration file, modify it as necessary, and then drag and drop all datasets to be processed into the appropriate box on the website to analyze data using. No additional user input is required: In order to provide highly scalable and entirely reproducible computational processing via IaC, NeuroCAAS first detects the submission event (dataset and configuration file submission), after which it hires a job manager to programatically create and manage all infrastructure and autonomously carry out analysis (Figure 3). Following the delivery of analysis results to the user (which may include live status logs and a detailed explanation of analysis parameters), the supplied infrastructure is then automatically destroyed once data processing is complete.
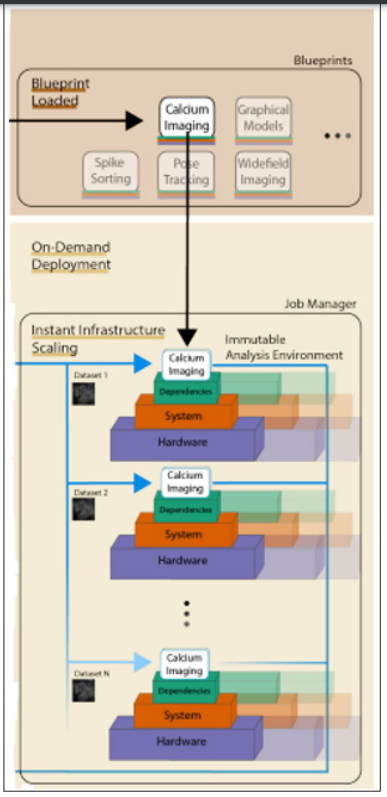
Figure 3: NeuroCAAS Workflow [27].
By utilizing fully virtualized infrastructure, the exact infrastructure resources used by the developer of an analysis are becoming available to all users. Additionally, this access is attained without limiting developers’ freedom to distribute resources according to their needs.
However, NeuroCAAS doesn’t allow users to deploy custom IAAC pipelines in their private cloud environment. Only the published pipelines on the platform can be utilized for data processing.
Whereas in this paper we wanted to enable all researchers to have their custom IAC code either shared by pipeline developers or researchers. Consider IAAC code just as a docker file which enables researchers to fire up a computational environment on the cloud and run the analysis pipeline.
Among main concerns, the following is distinguished: cloud computing basics and ramp up learning; billing and cost overhead which can easily balloon; addressable by built-in guard rails. One of the main challenges of cloud computing cost management is the complexity and variability of the cloud pricing and billing models. Moreover, overprovisioning of containerized applications adds to cloud costs [28, 29]. There is quite a potential challenge of idle resources, over provisioning, and orphaned resources. These unnecessary costs can quickly inflate cloud bills.
While IaC provides a great way to keep track of infrastructure changes and monitor things like infrastructure drift, maintaining an IaC setup becomes a challenge in itself once it reaches a certain scale. Also, vendor-agnostic IaC tools (such as Terraform) often lag behind on features compared to vendor-specific products. This is because tool vendors need to update providers to fully embrace new cloud features being released at an ever-increasing pace.
In addition, Infrastructure-as-Code needs additional tools such as a configuration and automation/organization management system, which can cause errors in the system [30]. Any bugs can quickly propagate across servers, especially where there is extensive automation, so version control and extensive pre-testing is very important.
With this perspective, IaaC definitely has a lot to cater to researchers without going into the complex stages of infrastructure deployment which can be challenging. Overall, the concerns and drawbacks, whatever they are, can be gradually overcome due to building a sustainable and open-source user and developer community around appropriate tools and platforms [31, 32].
In conclusion, the integration of Cloud IaaS in the field of neuroinformatics research offers a myriad of advantages. These benefits encompass the empowerment of end users, enabling dynamic scalability, providing access to specialized hardware, and capitalizing on the advantages of cloud-based technology, such as simplified deployment, flexible user management, robust data governance, and the use of specialized frameworks. By embracing these advantages and implementing automation for IaaS deployment to address any associated challenges, researchers can significantly enhance their neuroscientific investigations, streamline computational workflows, and achieve more impactful results. It is possible that, similar to the mainstream adoption of container image sharing, sharing IaaS templates alongside analysis pipelines could play a pivotal role in democratizing neuroinformatics and enhancing reproducibility in the field.
