Author(s): Mohitosh Biswas
Variability of ACE2 expression encoded by the ACE2 gene may be important for susceptibility and clinical outcomes of SARS-CoV-2 infection. This study was aimed to identify potential single nucleotide polymorphisms (SNPs) of ACE2 relevant to SARS-CoV-2 infection and predictively assigned risk phenotypes. Allele and genotype information of rs2285666 SNP of ACE2 was obtained from the 1000 Genomes project Phase III in line with Fort Lauderdale principles. About 16 SNPs of ACE2 as potential venture for susceptibility to SARS-CoV-2 infection was identified from the literature. Predicted high-risk phenotypes of ACE2 expressor due to carrying rs2285666 SNP of ACE2 was highly prevalent in East Asia (40.7%; 95% CI 36%-45%), followed by South Asia (36.8%; 95% CI 33%-41%), America (22.8%; 95% CI 18%-27%), Europe (14.5%; 95% CI 11%-18%) and Africa (12.3%; 95% CI 10%-15%), respectively. In total, ~25% of the world populations were predictively identified as being at high-risk for SARS-CoV-2 infection due to carrying rs2285666 ACE2 genetic polymorphism. Identification of high-risk phenotypes for SARS-CoV-2 infection through screening of ACE2 genetic polymorphisms may be valuable for SARS-CoV-2-related COVID-19 prevention and treatment in the population. Customized DNA microarray techniques or next generation sequencing may holistically advance this newly evolving research area of infection genetics.
Angiotensin-converting enzyme 2 (ACE2) encoded by the ACE2 gene has impacts on renin-angiotensin-aldosterone system (RAAS) and regulating cardiovascular effects. The ACE2 gene is highly polymorphic and many of the single nucleotide polymorphism (SNP) of this gene had been established as a risk factor for developing cardiovascular diseases [1,2]. Contrastingly, ACE2 has been proved to be the potential receptor for the severe acute respiratory syndrome corona virus (SARS-CoV) that outbreak in 2003 worldwide and also for the recent more devastating highly genomic homologous of the SARS-CoV called SARS-CoV-2 [3-6]. The SARS-CoV-2 associated disease called coronavirus disease-2019 (COVID-19) is causing huge morbidity, mortality and socio-economic burdens throughout the world. Recent studies and analyses indicate that ACE2 may be the main receptor for SARS-CoV-2 host entry through binding with the viral spike (S) protein [5,7,8]. A positive correlation of ACE2 expression and the in vitro infection of SARS-CoV was found in previous studies [9,10]. Therefore, the variability and magnitude of ACE2 expression in different human tissues governed by the genetic variants of ACE2 might be of critical for the susceptibility, symptoms and outcome of SARS-CoV-2 infection. As evidenced elsewhere, some patients may have higher expression of ACE2 as found in recent single-cell RNA-sequencing (RNA-seq) analysis [11,12]. After adjusting confounders, a very recent study elucidated that ACE2 gene expression in nasal epithelium was lowest in younger children and increased with age [13]. This reflects that unrevealed genetic polymorphisms of ACE2 might contribute this expression variability
However, the genetic polymorphisms of ACE2 positively correlated with the SARS-CoV-2 infection in different populations is still largely unknown. Therefore, the SNPs of ACE2 associated with increased risk for SARS-CoV-2 infection need to be explored at the population level. If it appears in the literature that any SNP was positively associated with increased risk for either SARSCoV-2 or SARS-CoV infection, then these polymorphisms may need to investigate among different populations to predict the risk phenotypes vulnerable to SARS-CoV-2 infection.
This study was designed to identify relevant SNPs of ACE2 that may associated with increased risk for SARS-CoV-2/SARS-CoV infection. The SNPs of ACE2 of interest was then explored in the world population using 1000 Genomes genomic data to predict risk phenotypes susceptible to SARS-CoV-2 infection.
Literature was searched in PubMed, Cochrane library, Scinapse and Google Scholar to identify studies that had either assessed the SNPs of ACE2 gene with SARS-CoV-2/SARS-CoV infection or suggested the SNPs that could possibly regulate ACE2 expression in different human tissues. Then make a list of these SNPs of ACE2 considering as potential candidate for investigating the genetic effects in future studies. Since it appeared in the literature that only rs2285666 SNP of ACE2 was found to be positively associated with the increased risk for SARS-CoV infection and suggesting that patients carrying this variant may had higher expression of ACE2 [14]. As SARS-CoV-2 has highly comparable genomic analogue properties with SARSCoV and no study has investigated the genetic effects of any specific SNP of ACE2 in SARS-CoV-2 infection, therefore, present study considered the rs2285666 SNP as potential candidate for exploring in the world population. Allele and genotype information of rs2285666 of ACE2 was obtained from the 1000 Genomes project Phase III in line with Fort Lauderdale principles. Participants carrying two copy of the rs2285666 SNP of ACE2 were predicted to high expressor of ACE2 and were potentially considered as high-risk phenotypes. In a comparative fashion, participants carrying one copy of the rs2285666 SNP of ACE2 were predicted to medium expressor of ACE2 and were potentially considered as medium-risk phenotypes. However, participants carrying no mutation of the rs2285666 SNP of ACE2 were predicted to normal expressor of ACE2 and were potentially considered as no-risk/normal phenotypes.
From the literature search, it was found that 16 SNPs (rs2285666; rs4646140; rs4646165; rs4646174; rs2301692; rs2301693; rs2074192; rs4646127; rs112171234; rs2158082; rs5936011; rs6629110; rs5936029; rs714205; rs4646142; rs2106809) with allele frequency greater than 1% of ACE2 may regulate ACE2 expression in different human tissues and may potentially interfere with SARS-CoV-2/SARS-CoV infection [3,14,15]. No study was identified in the literature to date that had assessed the association of these SNPs in the SARS-CoV-2 infection in COVID-19 patients. However, only six SNPs e.g. rs2285666; rs4646140; rs4646165; rs4646174; rs2301692; rs2301693 of ACE2 was found to investigate with SARS-CoV infection [14]. Among these, only rs2285666 SNP of ACE2 was found considerably high in SARS patients and indicated that 15 patients out of 20 SARS cases caused by SARS-CoV were carried minor allele and 32 out of 57 contact persons also carried this mutation. The findings of this study fueled that this SNP might also involve in the variability of SARS-CoV-2 infection in patients with COVID-19.
Using rs2285666 ACE2 genomic data from 1000 Genomes project, it was found that the variant allele was prevalent in 35.0% (95% CI 33%-37%) of the total 26 populations participated in this genomic project. It was also found that variant allele was prevalent highest in East Asia (53.7%; 95% CI 50%-57%) followed by South Asia (47.9%; 95% CI 44%-52%), America (33.6%; 95% CI 30%-38%), Europe (23.5%; 95% CI 20%-27%) and Africa (21.1%; 95% CI 19%-24%), respectively as shown in Figure 1.
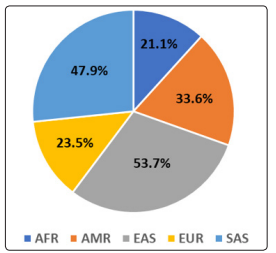
Figure 1: Predicted prevalence of rs2285666 ACE2 risk allele in different population participated in 1000 Genomes project. Here SAS: South Asian, EAS: East Asian, EUR: European, AMR: Ad.Mix American, AFR: African.
As described in Method section, high-risk phenotypes were prevalent in 24.7% (95% CI 23%-26%) of the total 26 populations participated in the 1000 Genomes project. It was also found that high-risk phenotypes were prevalent highest in East Asia (40.7%; 95% CI 36%-45%) followed by South Asia (36.8%; 95% CI 33%- 41%), America (22.8%; 95% CI 18%-27%), Europe (14.5%; 95% CI 11%-18%) and Africa (12.3%; 95% CI 10%-15%), respectively as shown in Figure 2. Interestingly, it was found that high-risk phenotypes due to carrying two copies of rs2285666 SNP of ACE2 were more of male participants predicted to representing high expressor of ACE2 than females (17.3%; 95% CI 16%-19% vs 7.3%; 95% CI 6%-8%).
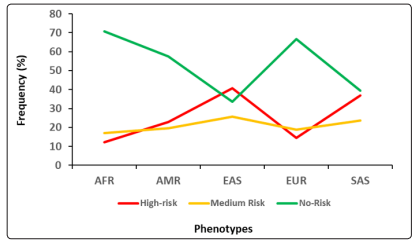
Figure 2: Predicted prevalence of phenotypes associated with rs2285666 ACE2 genetic polymorphism in different population participated in 1000 Genomes project. Here, SAS: South Asian, EAS: East Asian, EUR: European, AMR: Ad.Mix American, AFR: African.
The results of this study hypothesized that ~25% of the world population were susceptible to SARS-CoV-2 infection due to carrying just rs2285666 ACE2 genetic polymorphism and the rest ¼ of the world population may not be affected by this pandemic virus. The findings of this study warranted future clinical studies to prove experimentally this hypothesis.
In the COVID-19 pandemic, effective strategy to identify risk phenotypes of SARS-CoV-2 infection is of utmost importance to reduce huge economic, health and social burden of the world. Although developing herd immunity by posing the wider community to SARS-CoV-2 are getting most debatable approach in the current era to combat COVID-19, however, it should bean effective approach especially by inducing a greater health risk for the wider community. Instead, personalised genetic approach which can categorize the population as being as either high-risk or no-risk at all for susceptibility to SARS-CoV-2 infection may be considerably an effective strategy to screen general population.
For example, in this study just considering one SNP (rs2285666) of ACE2, about 1/4th of the world populations was predictively identified as being at high-risk phenotypes susceptible to SARSCoV-2 infection due to carrying rs2285666 ACE2 genetic polymorphism. It implies that majority of the world population may not being considered at risk for infected with SARS-CoV-2. For example, ~23% of the Americans were predictively considered as high-risk phenotypes for susceptibility to SARS-CoV-2 infection in this study. This prevalence may reduce considerably when it would be possible to taking into more SNPs of ACE2 as identified in this study in patients with COVID-19 under real world investigation with customized DNA microarray techniques. This is in line with the findings of recent studies indicating that only ~10% to 16% of the total COVID-19 suspected population screened for SARS-CoV-2 infection were found test positivity, although all the suspected patients were recruited from almost the same socio-economic conditions and had predisposed to similar health risk conditions[16-18]. This reflects that many patients inheriting some natural defenses and were contributing them not to be affected. The findings of the current study hypothesized that people inheriting rs2285666 ACE2 genetic polymorphism responsible for high expression of ACE2 were highly vulnerable to SARS-CoV-2 infection compared to those not inheriting this genetic polymorphism.
If this ‘proof-of-concept’ can be confirmed in future clinical studies with considering other potential SNPs identified in this study along with rs2285666 SNP of ACE2 relevant to SARS-CoV-2 infection, then it would be possible to identify risk phenotypes only for small proportion of people inheriting these polymorphisms and may implement effective interventions for susceptibility and severity of COVID-19 in these risk phenotypes. However, majority of the world population may not be affected by this pandemic virus due to not carrying the notorious genetic polymorphisms determined by the genetic screening process. Customized DNA microarray techniques or next generation sequencing may holistically advance this newly evolving research area of infection genetics. In addition, establishment of correlation between ACE2 expression governed by the genetic polymorphisms of ACE2 gene and SARS-CoV-2 infection in clinical studies may further greatly advance COVID-19 progression, treatments strategy and epidemiologic features. For example, a recent study found that COVID-19 patients with basic heart failure disease showed increased ACE2 expression at both mRNA and protein levels, indicating these patients may have higher risk of heart attack and critically ill condition if infected by this pandemic SARS-CoV-2 virus [2].
In this study, the rs228566 SNP of ACE2 was highly prevalent in males than females which may reveals that males may have more chance to get SARS-CoV-2 infection than females. This finding is consistent with many recent studies assessed SARSCoV-2 infection in COVID-19 patients [19,20]. This may further explain why mortality was significantly higher in male patients than females as observed from the international published data [20-22] which may be partly because of increased genetic mutation of ACE2 allowing males to be potentially high ACE2 expressor and rendering them to more risk for SARS-CoV-2 infection and poor clinical outcomes as well.
Linear regression with ACE2 gene expression in nasal epithelium as the dependent variable and age group as the independent variable, a recent study [13] showed that compared with younger children, ACE2 gene expression was significantly higher in older children (P = .01), young adults (P < .001) and adults (P = .001) which may partly able to explain why mortality was significantly higher in older patients as observed from the international published data [20,22,23].
The ACE2 is highly expressed in epithelial cells of the lung, intestine, kidney, and blood vessels which is predominantly upregulated in patients with diabetes, hypertension, kidney or liver diseases [11,24,25] and it has reported that comorbidities were significantly aggravating mortality in SARS-CoV-2 infection [20- 23], which again emphasized the considerations of ACE2 genetic polymorphisms regulating expression of ACE2 in COVID-19 patients in relation to disease progression and severe clinical manifestations.
As this is a predictive study and the ACE2 expression due to rs2285666 ACE2 genetic polymorphism was not measured in patients with SARS-CoV-2 infection, therefore, the findings of this study may vary with the real-world clinical investigation and therefore may not be immediately applicable. However, the design and framework of this study may be considered as novel to implement personalised approach to effectively manage unprecedented health situations due to COVID-19 pandemic.
Approximately 25% of the world population participated in the 1000 Genomes project was predictively identified as being at high-risk for SARS-CoV-2 infection due to carrying rs2285666 ACE2 genetic polymorphism. Personalised screening of ACE2 genetic polymorphism regulating ACE2 expression may be an effective strategy to combat COVID-19 pandemic.
Conflict of interest: None to declare.
Funding: No finding was available for this research.
Data availability: All data used in this study is available upon request.
