Author(s): Chito C Ekwealor, Amarachi F Ibeanu, Chikodili G Anaukwu*, Vivian N Anakwenze, Tobechukwu MC Ajogwu and Josephine C Ohuche
Serratia marcescens produces secondary metabolites, which are bioactive chemicals generated during its non-essential metabolic activities. These secondary metabolites are renowned for their antibacterial, antifungal, and anticancer capabilities. This work analyzed the chemical composition and antibacterial properties of the secondary metabolite of Serratia marcescens isolated from the local environment. Serratia marcescens was isolated using standard microbiological techniques and identified based on morphological and biochemical characteristics. Gas chromatographymass spectrometric analysis revealed the presence of eighteen (18) bioactive compounds, comprising oleic acid as the most abundant compound (21.9013 %), 9,12-Octadecadien-1-ol, (Z, Z)- (11.9854 %), and other compounds with less than 6 % abundance. The metabolite has antibacterial activities against Escherichia coli, Pseudomonas aeruginosa, Proteus vulgaris, and Bacillus subtilis. Antibacterial activity was higher against Bacillus subtilis (42.00±2.65 mm) and Proteus vulgaris (32.67±1.15 mm), and least with Pseudomonas aeruginosa (10.00±2.00 mm) at 400 mg metabolite concentration. The antibacterial effect decreased with a decrease in the concentration of the metabolite. The results show that Serratia marcescens exhibits potential as an antibacterial agent within the pharmaceutical and food sectors.
Serratia, a genus of Gram-negative bacteria, are known for their diverse biological activities, including antimicrobial agents, enzyme inhibitors, anti-tumor agents, immunotherapeutic agents, and plant growth stimulators [1-5]. They produce valuable organic compounds such as organic acids, biosurfactants, vitamins, lipids, pheromones, pigments, amino acids, cholesterol-lowing agents, symbiosis, receptor antagonists and agonists, biofuel, and food additives [3,6,7]. These metabolites are produced during the late growth phase (idiophase) of the microorganism when the exhaustion of a key nutrient source narrows growth [8].
Serratia species can grow on solid media at temperatures ranging from 20°C to 37°C and in liquid media at 5°C to 40°C with optimum pH values of 5 - 9. Some strains of Serratia have an extremely species-specific secretion system (type VI) also known as T6SS which enables the production of broad-spectrum bioactive compounds. This system helps the production of antibacterial toxins and self-protecting bacteriophage-contained proteins that add to virulence against competitors [9]. The production of bioactive secondary metabolites in Serratia is ascribable to Quorum Sensing (QS) [10]. There are various studies about the antimicrobial metabolites of Serratia marcescens, for example, the culture supernatant of Serratia marcescens 2170 has potent cytotoxic activity against cancer cell lines [11]. There have been reports of Serratia sp. strains ATCC 39006 producing broad-spectrum β-lactam antibiotic carbapenem and release of haemolysin by S. marcescens strain 274 and strain 39006 [12].
Secondary metabolites are classified into five classes, namely alkaloids, terpenoids and steroids, fatty acid-derived substances and polyketides, enzyme cofactors and nonribosomal polypeptides [13]. The alkaloids are amine groups, and the terpenoids and steroids are biologically synthesized from isopentenyl diphosphate. The fatty acid-derived substances and polyketides are biosynthesized from acyl precursors such as acetyl CoA, methylmalonyl CoA and propionyl CoA while the nonribosomal polypeptides are amino acid derivatives synthesized without direct RNA transcription. The nonprotein low-molecular enzyme components are the enzyme cofactors [14].
Secondary metabolites are controlled by peculiar regulatory mechanisms such as induction, carbon catabolic regulation, and feedback regulation [15]. They are not essential for the vegetative growth of producing organisms but are considered differentiation compounds conferring adaptive roles, such as acting as a defense weapon against other microorganisms, signaling molecules in ecological interaction, symbiosis, metal transport, competition, bioremediation, reproductive agent, and interference in spore formation and germination [13]. These bioactive metabolite compounds act as valuable resources for biotechnological applications, specifically for pharmaceuticals, nutraceuticals, and cosmetic industries.
This study aimed to profile the constituents of the secondary metabolites of Serratia marcescens and assess their antibacterial activity against clinical pathogens.
A total of 45 soil samples were collected from different farmlands, gardens, and flower beds around Nnamdi Azikiwe University, Awka. The Nnamdi Azikiwe University Awka campus is located on the Enugu-Onitsha Expressway in Awka, Anambra, Nigeria. The university is located in Southeast Nigeria, precisely between latitude 6.245° to 6.283° N and longitude 7.115° to 7.121° E. Replicate samples were obtained by random sampling at the sites at a depth of 5 cm from the surface using a sterile auger. The samples were labelled according to the sites of collection. The samples were then transported in sterile zip-lock polyethene bags in ice packs to the Department of Applied Microbiology and Brewing Laboratory.
One gram of each soil sample was introduced into 10 mL of distilled water. The suspension was serially diluted 10-fold, and 100μl of each soil suspension was placed on Glucose Yeast extract Calcium carbonate (GYC) medium for the isolation of Serratia species, composed of 5% glucose, 1.0% yeast extract, 2.0 % calcium carbonate and 1.5% agar, pH 5.94. The inoculated plates were incubated at 30?C for 24 hours. Developed colonies were subcultured on sterile GYC agar plates and further stored on nutrient agar slant at 40C [16].
Several biochemical tests and 16S rRNA sequencing were performed for the identification of the isolates. The biochemical tests carried out were gram reaction, catalase test, oxidase test, sugar fermentation test, methyl red test, and Voges-Proskauer test.
The 24-hour-old culture of the test isolate was smeared onto a grease-free slide using a sterile inoculating loop. A small amount (2 - 3 drops) of hydrogen peroxide solution was applied to the smear. The presence of gas bubbles on the surface of the slide indicated a positive result [17].
A solution of 0.1 g of oxidase reagent was prepared by dissolving it in 1 ml of sterile water. A sterile filter paper was immersed in the solution using sterile forceps and then left to dry. A small amount of the 24-hour-old culture of the isolate was applied onto the filter paper. The formation of purple colour signifies a positive oxidase test [17].
The sugars tested were lactose, glucose, sucrose, mannitol, galactose, D-xylose and myo-inositol. A loopful of the 24-hour- old culture of the test isolate was inoculated into a sterile sugar solution containing 1% sugar solution, 1% peptone water and bromothymol blue indicator in a test tube containing an inverted Durham tube. The tube was incubated for 24 hours, the colour change from blue to yellow indicated positive sugar fermentation, and the presence of bubbles at the tip of the Durham tube indicated gas production [17].
A 24-hour-old culture of the test isolate will be inoculated into 5 mL of glucose–phosphate peptone water and incubated for 24 hours at 37°C. Thereafter, 3 drops of methyl red indicator will be added to the culture broth. The production of a reddish color upon adding the indicator will signify a positive result, while a yellowish color will denote a negative result [17].
A 24 hr old culture of the test isolate will be inoculated into 5 mL of glucose–phosphate peptone water and incubated for 24 hours at 37°C. Thereafter, 5 drops of potassium hydroxide (KOH) will be added. The tubes will be shaken at intervals to ensure maximum aeration. The development of red color within 30 Sec and 60 Sec will indicate a positive Voges-Proskauer result, while the absence of red color will show a negative VP result [17].
The genomic DNA (16S rRNA) was isolated using the ZR bacterial DNA miniprep kit from Zymo Research. The DNA was subjected to polymerase chain reaction (PCR) amplification using Taq 2X Master Mix obtained from New England Biolabs (M0270). The primers utilized were the forward primer (27F: AGAGTTTGATCMTGGCTCAG) and the reverse primer (1525R: AAGGAGGTGWTCCARCCGCA). The cycling conditions for amplifying the 16S rRNA were as follows: initial denaturation at 94?C for 5 minutes, followed by 36 cycles of denaturation at 94?C for 30 seconds, annealing at 56?C for 30 seconds, and elongation at 72?C for 45 seconds. Afterwards, do a last elongation step at a temperature of 72?C for 7 minutes, and then maintain the temperature at 10?C. The DNA was subjected to electrophoresis on an agarose gel and observed using a UV transilluminator. The amplified fragments underwent sequencing using a Genetic Analyzer 3130xl sequencer from Applied Biosystems, following the instructions provided by the manufacturer. The sequencing kit utilized was the BigDye Terminator v3.1 cycle sequencing kit. The genetic study was conducted using Bio-Edit software and MEGA The evolutionary history was determined using the Neighbor- joining method, and the evolutionary distances were calculated using the maximum Composite Likelihood approach [18,19].
Secondary Metabolite Production by Submerged Fermentation A loopful of Serratia marcescens was introduced into 10 ml sterile distilled water in a test tube and standardized by referring to 0.5 Mc Farland standard. This served as the seed inoculum.
A submerged fermentation method was adopted for the release of metabolites by Serratia marcescens. De Man Rogosa and Sharpe (MRS) broth medium supplemented with finger millet was used as the fermentation medium. A 50 ml sterile fermentation medium in a 100 ml Erlenmeyer flask was inoculated with 1 ml of seed inoculum (1.2 × 106 cells/ml). The inoculated flasks were incubated for 14 days at 30 °C in a rotary shaker at 150 rpm. The experiment was carried out in triplicate and an uninoculated flask served as control. At the end of the incubation period, 30 ml of sterile distilled water was added to the flasks and followed by agitation for 5 minutes at 150 rpm. The fermentation broth was then allowed to stand for 24 hours at 25 0C and the content was centrifuged at 10,000 X g for 15 mins. The supernatant was separated using a pre-weighed Whatman No 1 filter paper, air- dried, weighed and used for further studies [20].
Ten grams of the filtrate was introduced in 10 ml of distilled water in a flask and subjected to solvent extraction to recover antibacterial metabolites in pure form. Ethyl acetate served as the solvent and was added to the suspension in a ratio of 1:1 v/v. The mixtures were kept for 15 minutes under periodic shaking. The ethyl acetate phase and aqueous phase were hoarded and concentrated by evaporation to near dryness in an oven.
Gas chromatography-mass spectroscopy was employed to identify the various compounds present in the secondary metabolite. A volume of one microliter of the extracted metabolite solution was introduced into a GC-MS instrument (Agilent Technology 5890) equipped with a split detector and Mass Spectrometer Detector. Helium was employed as the carrier gas with a consistent flow rate of 1 ml/min and an injection volume of 1 μl. The injector temperature was set at 250?C, while the ion-source temperature was maintained at 280?C. The overall duration of the GC process was 90.67 minutes. The identification of peaks was achieved by a combination of referencing their mass spectra and utilizing the NIST08 mass spectral database [21].
Clinical organisms, Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, Klebsiella pneumoniae, Proteus vulgaris and Bacillus subtilis were sourced from Clinical Diagnostic Laboratory NVRI Vom and used to test the antimicrobial activity of the antibiotic produced by Serratia marcescens. Filter paper disc diffusion technique was employed for the antibiotic susceptibility test. A loopful of the 24-hour-old culture of test organisms on a nutrient agar plate was standardized in 10 ml distilled water contained in a test tube to 0.5 Mc Farland standard. One millilitre of the standardised inoculum was spread inoculated onto sterile Mueller Hinton agar. The antibiotic disc was prepared by impregnating 0.01ml of the extracts onto a filter paper disc at varying concentrations of “400 mg, 200 mg, 100 mg, 50 mg, 25 mg and 12.5 mg.” The discs were placed onto the Mueller Hinton culture plate under sterile conditions. Growth inhibitions were measured after incubation for 24 h at 25 0C. Antibacterial activities were based on the zone of inhibition around the discs. The experiment was performed in triplicate and the zone of inhibition was recorded as mean±SD [20].
The analyses were conducted in triplicate, and the findings were recorded as mean±SD. The mean and standard deviation were calculated using Microsoft Excel 365. The analysis of ariance (ANOVA) of the data obtained during the antibacterial susceptibility test was done using SPSS version 20.
A variety of Serratia marcescens strains have been found in soil, freshwater lakes, and the ocean. These findings were reported by other researchers [20, 22 - 27]. The primary objective of this study was to isolate metabolite-producing Serratia marcescens from the local environment. The isolate’s basic biochemical features, as evaluated for preliminary identification of S. marcescens (Table 2), indicate that the organism is motile and positive for catalase enzyme production, as well as glucose, mannitol, galactose, D-xylose, and myo-inositol fermentation. However, it is negative for cytochrome oxidase enzyme production, Voges Proskauer and methyl red tests and lactose fermentation.
| Test | Result |
|---|---|
| Gram reaction | Gram-negative rod |
| Catalase | + |
| Oxidase | - |
| Motility | + |
| Citrate utilization | + |
| Methyl red | - |
| Voges Proskaeur | - |
| Sugar fermentation | |
| Lactose | - |
| Glucose | + |
| Mannitol | + |
| Sucrose | + |
| Galactose | + |
| D-xylose | + |
| Myo-inositol | + |
Extraction of genomic DNA from the isolate was performed, followed by sequencing and alignment using MUSCLE with default settings. The BLAST analysis determined that the sequences were Serratia marcescens and had a homology of 100% with strain KRED (NR_036886.1). The phylogenetic analysis revealed that several strains of S. marcescens, including strain Gol3 (NCBI accession number MT263018.1), strain P3 (NCBI accession number MZ477005.1), strain WES1 (NCBI accession number MN960115.1), and strain GMB S5 (NCBI accession number OR974900.1), demonstrated a pairwise similarity of 100%.
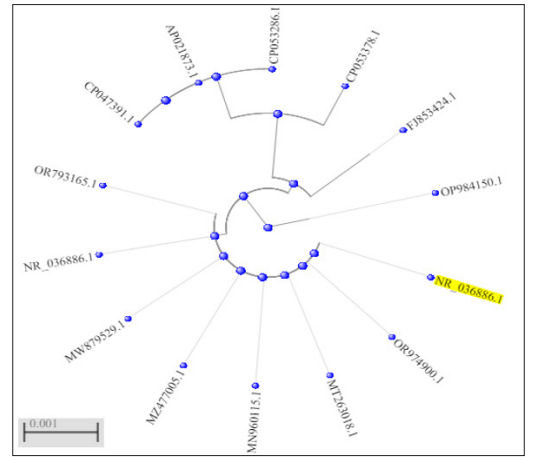
Figure 1: Phylogenetic Tree Showing Evolutionary Relatedness of the Isolate to other Strains of S. marcescens.
This work involved the profiling of the chemical components of the secondary metabolite using Gas Chromatography- Mass Spectrometry (GC-MS). The secondary metabolites of marcescens eluted between 12 and 36 minutes, resulting in the recovery of a total of 18 bioactive compounds (Figure 2). Compounds of aldehydes, fatty acids, β-Carotene, terpenoids, alcohol, and esters are hydrocarbons consisting of carbon rings ranging from 8 to 21. The component with the highest relative abundance was oleic acid, with a value of 21.9013%.
The list of aldehydes in Table 2 include 8-Hexadecenal; 14-methyl-, (Z)-; 9,17-Octadecadienal, (Z)-; 9-Octadecenal, (Z)-; 13-Octadecenal, (Z)-, and 9,12-Octadecadienal. 8-Hexadecenal, 14-methyl-, (Z)-, also referred to as (Z)-14-Methylhexadec- 8-enal, is a fatty aldehyde [28]. It has been documented as a primary component of the Scutellaria comosa plant, a medicinal herb employed for the traditional treatment of several illnesses in Uzbekistan [29,30]. It exhibits antimicrobial activity against multiple strains of bacteria and fungi, including Escherichia coli, Staphylococcus aureus, Enterococcus faecalis ATCC 29212, Pseudomonas aeruginosa, Candida albicans ATCC 90028, Candida parapsilosis ATCC 90018, Candida krusei ATCC 6258, and Fusarium oxysporum [31,32].
9,17-Octadecadienal, (Z) is a naturally occurring compound present in endophytic fungus and medicinal plants [33-36]. It has antioxidant and antibacterial properties [37]. The fatty aldehyde, 9-Octadecenal, (Z)-, is officially recognised as a flavoring agent by the FAO/WHO, with octadecenyl aldehyde as its record name [38]. Notably, the chemical has been isolated as a biologically active byproduct from an endophytic fungus, Paecilomyces sp (JN227071.1), which has antifungal properties against the disease- causing Rhizoctonia solani [39].
13-Octadecenal, (Z)-, is a chemical molecule classified as an alpha, beta-unsaturated aldehyde. It has been found in the leaf extract of Lindera setchuenensis and Bidens pilosa with antimicrobial properties and identified as a primary component of Citrus bergamia essential oil which has more potent antibacterial effects against Staphylococcus aureus ATCC6538 compared to Enterobacter ATCC 13048 [40-42]. Promising antibacterial efficacy against Staphylococcus aureus, Bacillus cereus, Bacillus subtilis, Escherichia coli, Salmonella typhimurium, Pseudomonas aeruginosa, Klebsiella pneumonia, and Candida albicans has been shown by the recovery of 9,12-Octadecadienal in endophytic Aspergillus species. Therapeutic plants such as Annona muricata seeds, Jacaranda cuspidifolia leaves and Clitoria ternatea L contain it as a bioactive component [43-45].
This work has identified 9,12-Octadecadien-1-ol, (Z,Z)-, also referred to as octadeca-9,12-dien-1-ol, as a secondary metabolite belonging to the alcohol class. This compound was found to be the highest detected compound in GC-MS analysis. It is found in most plant essential oils and has antibacterial, antioxidant, and anti-inflammatory characteristics [46,47]. Previous studies have documented it as a secondary metabolite. For example, Krishnaveni reported the presence of 9,12-Octadecadienal among the compounds recovered in their study [48]. Similarly, Rajput and Bithel observed a relative abundance of 0.54% of the compound in the GC-MS analysis of Hydnocarpus laurifolia [49]. 9,12-Octadecadien-1-ol, (Z,Z)- is a constituent of the endometabolite produced by honeybees, as well as from the seeds and seed oil of Millettia pinnata and the extract of Hydnocarpus laurifolia (Dennst) [49,50]. An investigation demonstrated the antibacterial, antibiofilm, antioxidant, and anticancer properties of a seed extract from Pongamia pinnata that contains 9,12-octadecadien-1-ol as a primary bioactive constituent [51]. Known for its skin conditioning properties, isopropyl linoleate is a widely used cosmetic component [52]. Significantly, it is reconstituted in an Indian herbal essential oil that has potent insecticidal properties against Aedes aegypti and Culex quinquefasciatus [53].
Esters recovered in this investigation include methyl-9,12- heptadecadienoate (2.02 %), E,E-10,12-Hexadecadien-1-ol acetate (5.21 %), Z,Z-10,12-Hexadecadien-1-ol acetate (0.68 %), 9-Octadecenoic acid (Z)-, 2-hydroxy-1-(hydroxymethyl) ethyl ester (0.08 %) and 9-Octadecenoic acid (Z)-, 2,3-dihydroxy-, propyl ester (0.10 %) (Table 2). These compounds are frequently present in medicinal plants that have demonstrated remarkable antibacterial properties. By way of illustration, methyl 9,12-heptadecadienoate was isolated from walnut oil, a compound commonly employed in pharmaceuticals [54]. Similarly, Z, Z-10,12-Hexadecadien-1-ol acetate was detected in the crude extract of Cucurbita moschata (pumpkin) seeds and Benincasa hispida [55]. 9-Octadecenoic acid (Z)-, 2-hydroxy-1-(hydroxymethyl) ethyl ester was identified as a constituent of Abrus precatorius L. with antibacterial, insecticidal, antiprotozoal, antiparasitic, anti-inflammatory, antioxidant, and immunomodulatory properties [56]. The moiety 9-Octadecenoic acid (Z)-, 2,3-dihydroxy-, propyl ester was isolated from the root of Rhazya stricta [57] and from the endophytic fungus Cunninghamella bigelovii. This compound has potential industrial applications in several sectors such as food, biological pharmacy, and chemical industry [58].
Oleic acid, cis-vaccenic acid, Z-4-Nonadecen-1-ol acetate – and 9,12-Octadecadienoyl chloride, (Z, Z)- are components of the fatty acid group. Oleic acid is an endogenous fatty acid with antibacterial properties that is used in several pharmaceutical formulations. It has been isolated as a medicinal compound from the intracellular extract of Glutamicibacter mysorens [59]. It is present in several extracts of biological products which are effective against microbial infections such as Moringa oleifera Lam. Leaves and seeds, Glycine max oil, Oenothera biennis L, and apricot seed oil [60-63]. A study conducted by Pushparaj et al. found that oleic acid improved antibiotic action against multidrug-resistant Pseudomonas aeruginosa strains [64]. Cis- vaccenic acid has been shown to exhibit antiviral, antioxidant, and anti-inflammatory characteristics. Dembitsky et al. have identified it as a prominent fatty acid component of the cellular slime mold, Dictyostelium discoideum [65]. This compound can be found in the leaves of Moringa oleifera and Solanum khasianum, as well as in the bark extract of Quercus leucotrichophora A. Camus Z-4-Nonadecen-1-ol acetate was recovered in ethanolic extract of fenugreek seed while 9,12-Octadecadienoyl chloride, (Z, Z)- was found in Amphora coffeaeformis extract and four wild edible macrofungi (Auricularia auricula-judge (Bull.) J. Schröt, Pleurotus ostreatus (Jacq. ex Fr.) P. Kumm, Pleurotus tuber-regium (Rumph. ex Fr.) and Schizophyllum commune. Furthermore, these macrofungi have antibacterial and antioxidant characteristics [66-69].
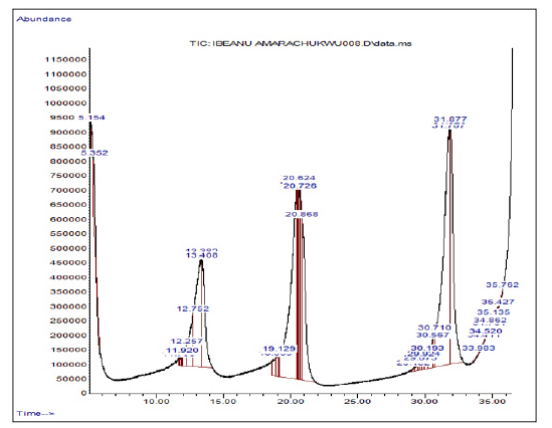
Figure 2: GC-MS Chromatogram of the Bioactive Compounds Present in the Secondary Metabolite of Serratia marcescens
| S/N | Retention Time (mins) | Compound | Relative Abundance (%) | Nature | Chemical formula |
|---|---|---|---|---|---|
| 1 | 5.154 | Oleic Acid | 21.9013 | Fatty acid | C18 H34 O2 |
| 2 | 31.787 | 9,12-Octadecadienoyl chloride, (Z,Z)- | 21.5821 | Fatty acid | C18H31ClO |
| 3 | 31.877 | 9,12-Octadecadien-1-ol, (Z,Z)- | 11.9854 | Fatty alcohol | C18 H34O |
| 4 | 5.352 | 8-Hexadecenal, 14-methyl-, (Z)- | 5.352 | Aldehyde | C17 H32O |
| 5 | 13.409 | E,E-10,12-Hexadecadien-1-ol acetate | 5.2143 | Fatty ester | C18 H32 O2 |
| 6 | 11.863 | 9,17-Octadecadienal, (Z)- | 4.7361 | Fatty aldehydes | C18 H32O |
| 7 | 12.752 | Isopropyl linoleate | 3.0308 | Ester | C21 H38 O2 |
| 8 | 11.706 | Methyl 9,12-heptadecadienoate | 2.0179 | Methyl esters | C18 H32 O2 |
| 9 | 20.513 | 13-Octadecenal, (Z)- | 1.7119 | Fatty aldehyde | C18 H34O |
| 10 | 18.869 | 9-Oxabicyclo[6.1.0]nonane | 1.687 | Oxabicyclic compound | C8 H14O |
| 11 | 30.567 | 9,12-Octadecadienal | 1.4704 | Aldehydes | C18 H32O |
| 12 | 12.257 | 9-Octadecenal, (Z)- | 0.9368 | Aldehyde | C18 H34O |
| 13 | 19.072 | cis-Vaccenic acid | 0.6964 | Fatty acid | C18 H34 O2 |
| 14 | 30.71 | Z,Z-10,12-Hexadecadien-1-ol acetate | 0.6773 | Ester | C18 H32 O2 |
| 15 | 34.764 | 9-Octadecenoic acid (Z)-, 2,3-dihydroxy-, propyl ester | 0.1015 | Ester | C21 H40 O44 |
| 16 | 29.152 | 9-Oxabicyclo[6.1.0]nonane, cis- | 0.0947 | Terpenoid | C8 H14O |
| 17 | 35.135 | 9-Octadecenoic acid (Z,Z)-, 2-hydroxy-1- (hydroxymethyl) ethyl ester | 0.0771 | Ester | C21 H38 O44 |
| 18 | 34.411 | Z-4-Nonadecen-1-ol acetate | 0.0632 | Fatty acid | C21 H40 O2 |
Experiments were conducted to evaluate the antibacterial efficacy of the secondary metabolite against Escherichia coli, Pseudomonas aeruginosa, Proteus vulgaris, and Bacillus subtilis. As seen in Table 3, all the test organisms exhibited susceptibility to the metabolite at the various concentrations examined. However, there were notably greater inhibitory effects observed at 400 mg. These findings suggest that the antibacterial efficacy of the metabolite is directly proportional to the concentration. A pictorial depiction of the experiment is shown in Figure 3. The susceptibility of the organisms to the metabolite exhibits substantial variation (P < 0.05). Antibacterial activity is considered strong when the zone of inhibition is greater than or equal to 20 mm, while moderate and weak activity is indicated by zones of inhibition of 10 mm and less than 10 mm, respectively [70]. Therefore, the secondary metabolite of marcescens in this work exhibited potent antibacterial action against P. vulgaris and B. subtilis at all doses, except for 12.5 mg. This result corroborates the report of other researchers. Karayildirim et al. reported the antibacterial activity of Serratia marcescens metabolite against Staphylococcus aureus with a 29.4 mm zone of inhibition [71]. The crude extracts of S. marcescens P1 and NP1 displayed broad-spectrum antimicrobial activity against clinical, food and environmental pathogens, such as multidrug-resistant Pseudomonas aeruginosa, methicillin-resistant Staphylococcus aureus and Cryptococcus neoformans [6]. Serratia marcescens could be potentially used as a broad-spectrum therapeutic agent against multidrug-resistant bacterial and fungal pathogens.
| Test Organism | Zone of Inhibition (mean±SD) in mm | |||||
|---|---|---|---|---|---|---|
| 400 mg | 200 mg | 100 mg | 50 mg | 25 mg | 12.5 mg | |
| Escherichia coli | 17.00±2.00 | 13.67±0.58 | 12.00±1.73 | 9.33±2.31 | 7.33±0.58 | 5.33±1.15 |
| Pseudomonas aeruginosa | 10.00±2.00 | 7.67±0.58 | 6.67±0.58 | 6.33±0.58 | 6.67±1.15 | 5.67±0.58 |
| Proteus vulgaris | 32.67±1.15 | 30.33±2.52 | 27.33±0.58 | 27.33±0.58 | 21.00±1.73 | 18.67±1.15 |
| Bacillus subtilis | 42.00±2.65 | 31.67±0.58 | 29.33±0.58 | 26.67±1.15 | 24.67±1.15 | 18.67±2.08 |
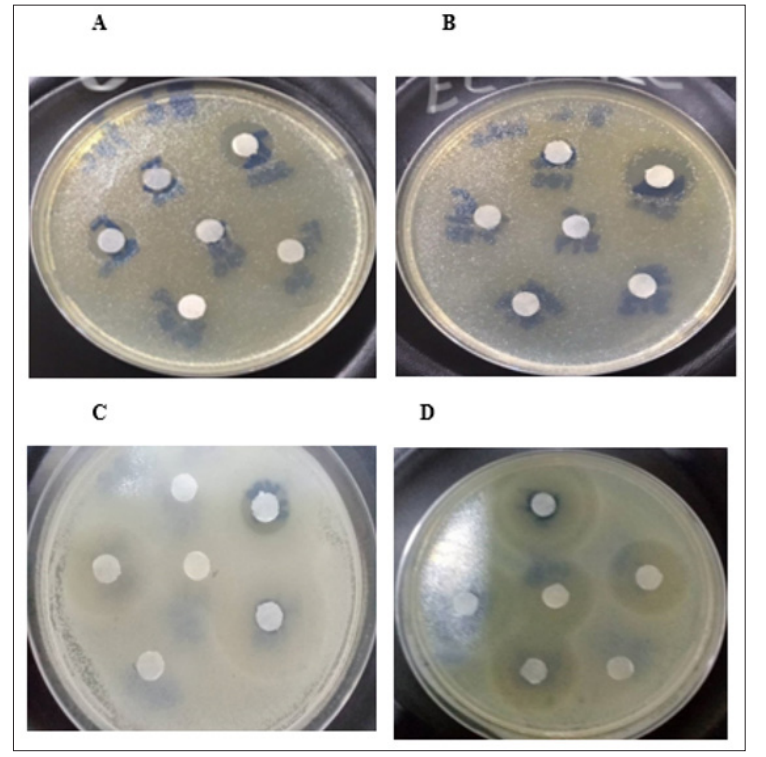
Figure 3: Pictorial Representation of the Antibacterial Susceptibility of the Test Organisms
where (A) E. Coli (B) P.aeruginosa (C) P. vulgaris and (D) Bacillus subtilis
The authors acknowledge the Ecology Laboratory of A.P. Leventis Ornithological Research Institute Lamingo, Plateau State, Nigeria and Mr Monday I Okpanachi, for providing laboratory space, guidance and resources for the research.
