Author(s): Prakash DSRS *, Murali Mohan S and Balaji Chandra Mouli J
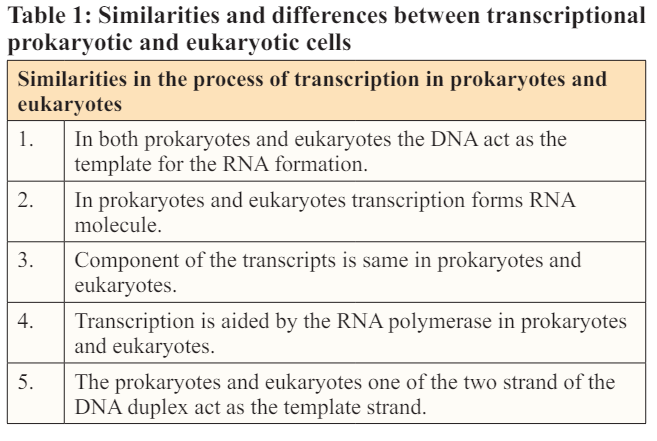
This article is totally based on the literature review about expression of gene in prokaryotic and eukaryotic cells. Gene is the part of the DNA that is responsible for the formation of the genotype and then a phenotype. Gene can only be expressed in case of formation of protein. The process of the gene expression is accomplished in the two steps; Transcription and translation. Transcription is the process of the formation of the mRNA. Transcription proceeds in the three steps. In initiation step the RNA polymerase moves on the unwind DNA strand until the promoter sequence is reached, next is the elongation step in the newly synthesized RNA strand elongates, the process terminates as the RNA polymerase realize the termination sequence. After the mRNA is formed it is released into the cytoplasm without any alterations in case of prokaryotes, but in case of eukaryotes post transcriptional modifications takes place which include capping, tailing and splicing. Translation is the process of the formation of protein from mRNA. Translation proceeds in the four steps in which the t RNA is charged, the initiation complex is formed with Met-tRNA at the P site start codon is recognized after binding of the ribosomal subunits to mRNA, in elongation step the peptide bond is formed and the polypeptide chain grows, stop codons enters at the A site and thus the process is terminated. Post translational modications takes place which the initiation amino acid methionine is removed.
Proteins are of great importance for both prokaryotes and eukaryotes, they are involved in various functions including repairing of the body tissues, controlling and coordinating various functions of the body [1]. Protein synthesis is carried upon by the body in two main steps transcription and translation which ae briefly described in the article below [2]. These two processes are similar in the prokaryotes and eukaryotes with the minor exceptions. Transcription involves the synthesis of the mRNA from the DNA, after synthesis the transcribed strand of the mRNA is immediately converted into the protein in case of prokaryotes, but in case of the eukaryotes mRNA undergoes post transcriptional modifications before being released into the cytoplasm as in eukaryotes the mRNA is at the risk of the degradation by the various enzymes that are absent in prokaryotes. Before post transcriptional modification the mRNA is nonfunctional and is considered immature. The post transcriptional modifications include three major steps; Capping, splicing and tailing. The two above mentioned processes must be coordinated and occur in time in order to meet protein requirement of the body [3].
Chromosomes are the structures that are present inside the cell. Chromosomes contain several genes. DNA consists of the several segments which are referred as gene [4]. Gene expression is known to be the process through which the information which is present on the genes is encoded to form a RNA molecule which further makes proteins or there could be the formation of the noncoding RNA molecules that does not form RNA but perform other important function in the body. Genes are not active as they are in their compact form. Factors which are involved in the opening and closing of the genes helps to access the genes [5]. Step of gene expression is very important in determining the location of the RNA molecules and thus the protein, and along with the location it also controls the amount of the RNA and proteins formed. The process of gene regulation is precisely controlled and very minute changes are made under unusual conditions. The products of RNA and the genes formed as a result also regulate the expression of the other genes.
The process of the gene expression is accomplished in two steps:
• Transcription
• Translation
Proteins are formed from the genetic information coded in DNA [6]. Transcription is the process of the formation of the mRNA from a specific DNA sequence. Those pieces of the DNA that are transcribed into RNA and are going to encode a protein are known as messenger RNA (mRNA).Other pieces of DNA which are duplicated into the RNA molecules and do not code for theprotein are known as the non-coding RNAs [7]. In the body numerous cells in a tissue has 10 times more the quantity of the coding mRNA as compare to the non-coding RNA (in specific cells depending upon the functionality this ratio might change).During the process of the transcription ,a sequence of the DNA is read by the RNA polymerase (transcription of nucleus encoded genes in eukaryotes is performed by RNA polymerase I, II, III,which produces the RNA strand which is complementary and antiparallel, this strand is known as primary transcript) [8 ].
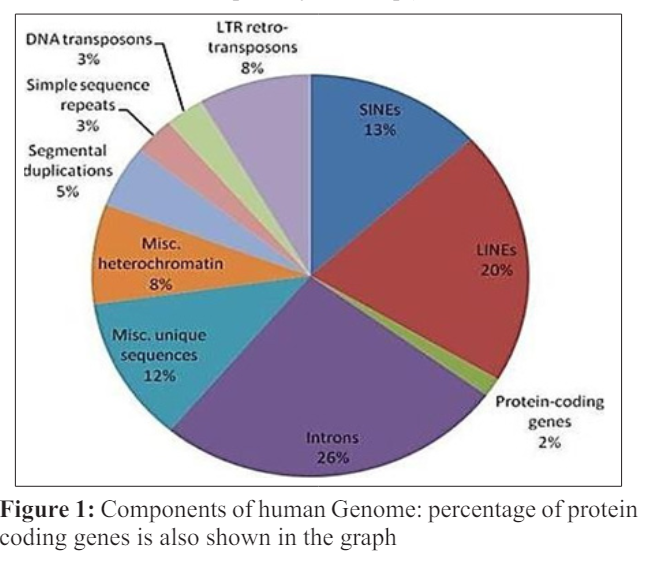
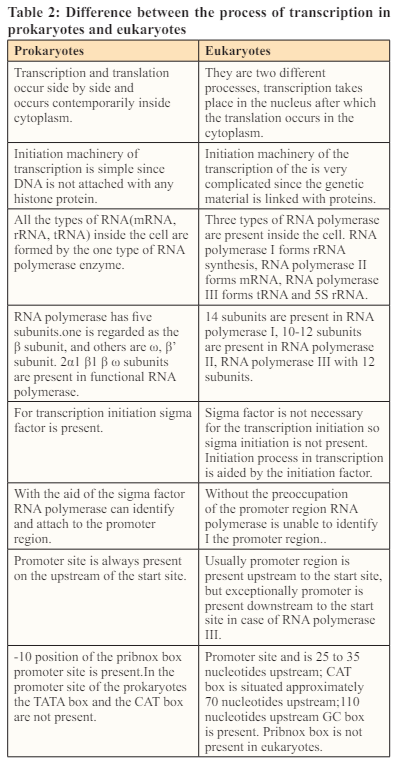
Transcription proceeds in the following step; 1) RNA polymerase attaches to the promoter DNA and also with the general transcription factors (SP1 is the very famous transcription factor, as a result the DNA strands are unwound and transcription bubble is formed [9]. 2) Inside the transcription bubble RNA polymerase segregates the two strands of the DNA helix, in this step the hydrogen bonds in the complementary strands of the DNA are broken down.3) RNA polymerase adds the nucleotides of the RNA which are opposite to the nucleotides sequence of the DNA strand. 4) With the help of the RNA polymerase backbone of the RNA sugar -phosphate is made to make an RNA strand. 5) RNA -DNA helix breaks because of the breaking of the hydrogen bonds in the helix, as a result the RNA strand is free. 6) If the cell consists of the nucleus as in case of eukaryotes, the RNA is further processed and this might include polyadenylation, capping and splicing. 7) Through the nuclear pore RNA might move to the cytoplasm or reside inside the nucleus.
The initial step in the transcription is the initiation in which the RNA polymerase binds to the 5’ end of the DNA at a specific sequence site which is known as promoter [10]. In the smaller organisms like bacteria, promoters are composed of the three basic elements whereas in eukaryotes there are up to seven elements comprising the promoter. In prokaryotes, most of the genes have a specific sequence known as the pribnox, containing a specific sequence TATAAT present 10 base pairs far from the site where transcription initiation takes place. It is not necessary that all the pribnox have this sequence of the nucleotides, but this sequence is the most frequent sequence found at almost every site .There are several alternative sequences of the nucleotides present in each box but resemble the original sequence of the pribnox closely. Several genes have the sequence TTGCCA at the 35 base position upstream the initiation site and some of the cells have an upstream element, which is rich in A-T of 40 to 60 nucleotides upstream that influence the rate of the transcription. In order to enhance the access to gene, the DNA double helix is unwound with the help of the holoenzyme. When the RNA polymerase “core enzyme” binds to another subunit which is known as sigma subunit, a holoenzyme is formed which unwind the double helix of the DNA. The sigma subunit guides RNA polymerase where to bind. Several different sigma factors are present which help in turning off and on of genes in various conditions by binding to different promoter.

Promoters in the eukaryotes are more complex as compare to the prokaryotes, because eukaryotes have three classes of the RNA polymerase that are capable of transcribing different sets of the genes. Promoters are the best transcriptional regulatory sequences in genome [11]. Many genes of the eukaryotes have an enhancer sequences which are found at a reasonably large distances from the sets of the gens they effect [12]. Gene activation is controlled by binding of the enhancer proteins with the activator proteins and changing the 3-D structure of DNA which is now capable of attracting RNA polymerase II, thus regulating transcription. As in the eukaryotes the DNA is tightly packed as chromatin, so the large number of specialized genes are required that will help to reach the template strand. Several cofactors are being used by the RNA polymerase of the eukaryotes. One cofactor is TFIID which effects the rate of the transcription and also determine the initiation site by recognizing the TATA box (the promoter sequence in eukaryotes.) Another one is TFIIB which recognize the specific sequences comprising 38 to 32 base pairs upstream.
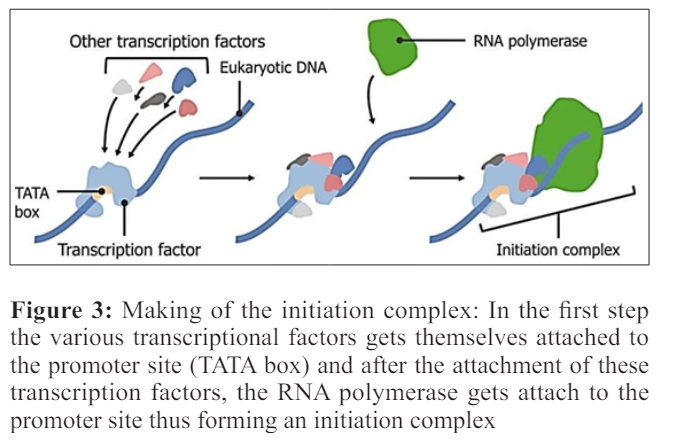
Enhancer sequences enhance the transcription of the genes. Enhancers were recognized 30 years ago while studying the regulation of SV40 virus early gene transcription [13]. Enhancers can be more or less thousand nucleotides far from the promoter with which they interact but looping of the DNA brought them into the close proximity. This looping mainly occurs as a consequence of the attractions between the proteins that bound to the enhancer and to the promoter. Those proteins which help in the process of the looping are known as the activators and those inhibit the process of the looping are known as the repressors. Transcription with the help of the polymerase I and polymerase III occurs in the same manner but the promoter sequences and the transcriptional activator proteins vary.
One of the two strands of the DNA (non-coding strands) is used as the template for the RNA formation. As the process of the transcription continues, template strand is traversed by the RNA polymerase and synthesize the RNA copy using the complementary base pairing of the template strand of the DNA. The transcription proceeds from 5’ to 3’so therefore copy of the RNA molecule from 5’ to 3’ direction which is the exact copy of the coding strand with the difference that the thymine are altered with uracil and the nucleotides are made of the ribose sugar rather than the deoxyribose sugar [ 14].
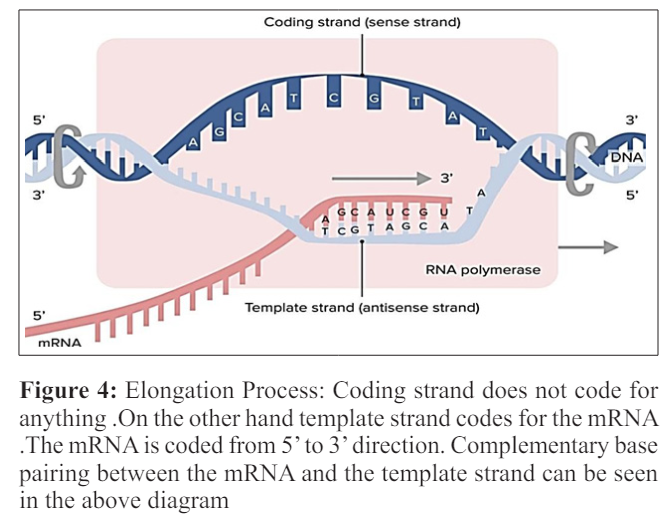
Multiple RNA polymerases can be involved in the transcription of the mRNA on a single template of the DNA so large number of the mRNA molecules can be formed from one copy of the gene. The rates of the elongation in the prokaryotes and the eukaryotes are about 10-100 nts\sec. During the process of the elongation in transcription in the eukaryotes the nucleosome acts as the major barrier. So in eukaryotes the pause that is induced by the nucleosome can be regulated by the transcription elongation factors such as TFIIS [15]. Proofreading step is also involved in the process of the elongation mechanism in which the incorrectly incorporated bases are eliminated .The short pauses in the process of the transcription allows the appropriate factors of RNA editing to bind.
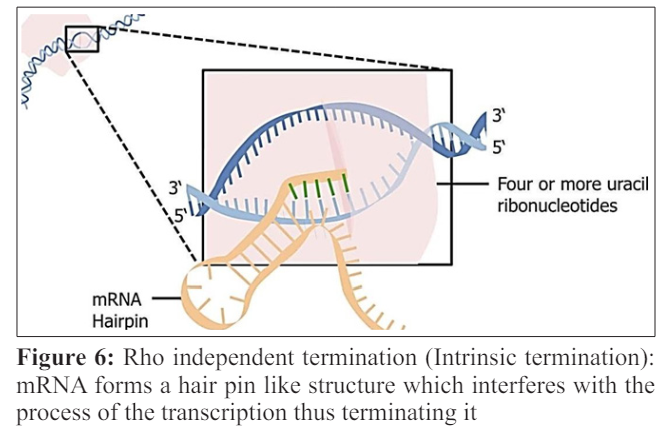
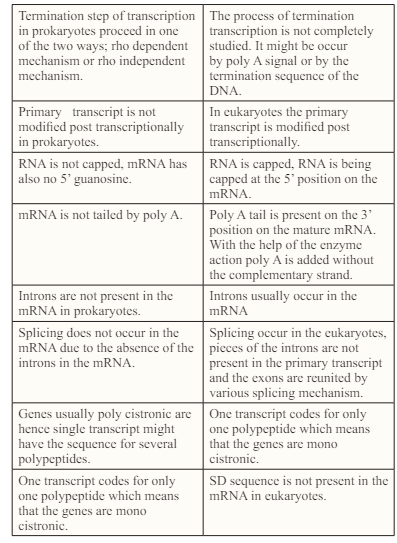
This step of the termination marks the end of the formation of the mRNA. This step proceeds in two ways ;Rho dependent termination and Rho independent termination. Before going into the details of the above mentioned two processes, one must understand what actually Rho is? “Rho factor is a hexagonal circular ring like shape helicase which brings to an end transcription in Escherichia coli [16]. Two recently discovered lattices of Rho in complex with nucleic acid shows the phenomenon of loading of this ring by mRNA and encircling it.” In Rho dependent transcription, the Rho factor gets attach to the transcript of the mRNA and use its enzyme which in this case is helicase function to move along the transcript towards the RNA polymerase. As the Rho reaches the polymerase it not only detaches the mRNA from genomic DNA but also detaches the RNA polymerase.
Rho structure is being studied for years and years, and the latest structure from the berger lab unite with the studies that shows the mechanism of hexameric helicase loading on the mRNA [17]. Two structures of the Rho are given out which fancy Rho’s binding to the nucleic acid, either with or withiut the utilization of ATP. In both cases, the hexamer of Rho is a broken circle which resembles a ‘lock ring’ in appearance.The gap in the ring is nearly 12 A wide; this width is enough to accept the strand of the mRNA in the center of the hexamer. With the help of the electron microscope, unfolded forms of the Rho have been seen.
The monomer unit of the Rho is composed of the two domains, each of the domain binds to the RNA. One is the amino- terminal domain ,which has 1° mRNA binding site and loads onto the target sequence of the mRNA (which is called as the rut sequence) .The carboxyl -terminal domain which has 2° m RNA binding site, attaches the mRNA for helicase or transferring activity. The mRNA which binds to the 1°site is not located at the periphery rather it is present at the middle of the ring, and directs the m RNA inward toward the central cavity (step1). Earlier studies under the electron microscope shows, that the grooved form of Rho was not seen in the existence of the nucleic acid, probably due to the closure of the ring. This study along with the newly discovered structure leads to the view that how mRNA is placed in the middle of the Rho. Through this opening m RNA is directed from 1° sites to the 2° sites .In case if the Rho is closed initially the m RNA cannot enter the ring of the Rho and will be attached proximal through the interaction between 1° site until the unfolded state if the Rho is presented .Thus one can conclude that the binding of the mRNA to the 1° sites promotes the unfolded form of the Rho .When the RNA binds to the 2° site, the ring shuts (step2) [17]. The unfolded form of the Rho, cannot begin the catalytic hydrolysis of the ATP .So the closure of the ring starts the ATP hydrolysis, therefore propels Rho in activity of RNA polymerase (step3) [18]. In the last step, Rho starts unwinding RNA from the genomic DNA and the RNA polymerase detaches (step4) [19, 20].
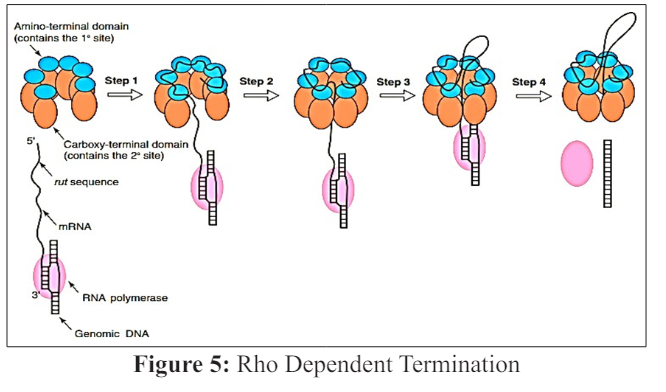
Aminoacyl -terminal domain of Rho factor (blue ) which has the site that binds with the 1° m RNA, attaches to the rut sequence present on the mRNA Rho factor is shown up as an unfolded ring but it might be in balance between the folded and the unfolded states (step 1).Carboxyl -terminal domain of Rho (orange) which has the site for binding the 2° m RNA, binds to the m RNA lower extreme from the rut and the hexagonal ring folds (step 2) .The carboxyl -terminal domain of the Rho propels itself along the m RNA in the 5’ to 3’ direction .The amino -terminal domain disengages itself from the rut during this sequence. (step3) Rho factor act as the helicase to pair ATP binding and hydrolyze the unwounded of the nucleic acid. As a result the mRNA is disengages from genomic DNA and RNA polymerase (pink)
Rho independent termination or intrinsic termination occurs when the machinery for transcription reaches the termination sequence [21]. Firstly, there comes a GC-rich palindrome; which causes the newly made RNA to form a hairpin like structure by forming base pair with itself. This hair pin loop disturbs the RNA-DNA complex. Next there comes the 4 or more uracil in the row; U-A bonds are weaker as compare to the G-C bonds .So these bonds are unable to keep the RNA on the DNA and the mRNA falls off.
After the process of the transcription the mRNA needs to be modified in the eukaryotes, so that it is prevented from the degradation and can safely traverse from the nucleus to the cytoplasm. There are three major steps involved in the process of the m RNA modification [22]. 1) Capping: is the process in which a methylated guanosine is added to the 5! end of the mRNA, the terminal G or A is methylated to form methyl guanosine or methyl adenocine. The terminal triphophate of hn RNA reacts with GTP under the influence of guanyl transferase before cap formation. It is followed by methylation.7 methylguanosine is added to the cap and this end becomes m7G CAP. it does not allow mRNA to bind to the to the RNA chains, protects m RNA from degradation, it helps the m RNA to travel from nucleus to the cytoplasm, facilitates the binding of the m RNA to the ribosomes to start translation. 2) Polyadenylation: a tail of the adenine molecule is added to the 3! terminus of hn RNA about 200 to 210 adenylate residues are added. It becomes poly A tail. Poly A polymerase catalyses this reaction. The Sequence is AAUAAA. The tail formation prevents the degradation of m RNA and increases the stability of the m RNA. 3) Splicing: The primary transcripts contain two types of nucleotide sequences. They are coding sequences called Exons and noncoding sequences called introns. The exons and introns are alternately arranged. During processing, the introns are exised from the primary RNA transcript. After the excision of introns, exons are joining together to form the final m RNA. Non coding introns are being spliced out by spliceosomes (enzymatic ribonucleoprotein complexes. Through differential splicing multiple different proteins can be formed from a single gene
“Translation is known as the formation of the protein from the mRNA.” The mRNA that is being transcribed in the first step of the gene expression needs to be translated in order to make a functional protein [23]. In translation the terms codon, anticodon and stop codon are frequently used. Codon is the specific sequence of the three nucleotides that is present on the mRNA and forms a genomic information that is going to code for a specific amino acid. There are total 64 codons out of which 61 code for the amino acid and the rest 3 codons are the stop codons. Start codon will mark the site where the translation will began, in eukaryotes the most common start codon is AUG which codes for methionine, in mammalian cells CUG codon can also be used as a start codon which codons for leucine and in prokaryotes start codons are GUG and UUG. There are three stop codons that are present in the genetic code and those are UAG, UAA, UGA and stops the process of the translation. These codons can also be termed as the non- sense codons or terminator codon as they are not going to code for any amino acid.
The machinery for translation includes; Ribosomes and transfer RNA (t RNA). Ribosomes are multifunctional complex which is composed of both RNA and proteins [24].Ribosomes are present in the cytoplasm. As soon as the mRNA is released into the cytoplasm ribosomes bind to it. It would not be wrong to say that the translation takes place inside the structures called ribosomes. Ribosomes not only catalyze the process of the translation but also connect various amino acids to make a polypeptide chain. The composition of the ribosomes vary from species to species which means that they might have different number of the ribosomal (rRNA) and polypeptides depending upon the organism and its protein requirement, however the general structure of the ribosome is same and is comparable to the bacteria. Ribosome is a complicated macromolecule that is made up of the larger and the smaller subunit [25].The small subunit binds the mRNA template while the larger subunit is responsible for binding of an t RNA (t RNA is the type of the molecule that is responsible for bringing different types of the amino acids, to the newly forming polypeptide chain).
Ribosomes have various slots for the transfer RNA (t RNA), t RNA is well explained in the next section. For now just remember that the ribosomes consist of the three different sites A (aminoacyl) site, P (peptidyl) site and E (exit) site [26]. Transfer RNA travels from the A site to the next site which is P to last region of the ribosome which is E site during the delivery of the amino acid during translation. Transfer RNA acts as the molecular bridge between mRNA and the amino acid. Transfer RNA is the unique sort of the molecule which matches codon on the mRNA with the specific amino acid it is coding for. Each tRNA consists of the group of three nucleotides known as anticodon. The anticodon present on the given t RNA can only attaches to either one or a some unique mRNA codons. The t RNA also has a amino acid especially, the one which is encoded by the codon to which the t RNA is going to bind.
There are various types of the t RNAs that are roaming around inside the cell, each consisting of its own anticodon and parallel amino acid [27]. The function of the anticodon is the recognition of the t RNA by amino acyl t RNA. There are 40 to 60 various types which depends upon different species. T RNAs bind to the ribosomes and thus the codons inside them and there they distribute amino acids for the formation of the polypeptide chain. Some t RNAs have the specialty of forming the base pairing with various codons. Base pair can be formed at the position third on the codon rather than a typical base pairing between A-U and G-C and this phenomenon is known as wobble [28]. Wobble pairing has its own rules rather than a typical base pairing .For example a G present in the anticodon can pair up with a C or U (but not with A or G) present on the third position. Rules like this make it sure that the codons are read flawlesssly despite a wobble. Wobble pairing makes it sure that the fewer t RNAs are covering all codons of the genetic code, it also makes it sure that the code is read accurately [29].
Aminoacyl-t RNA synthetase have the important task of determining the genetic code [30] .Three distinct synthetase enzyme are present for every amino acid, that will recognize only that specific amino acid and its t RNA .Once both the amino and the t RNA gets attached, the enzyme binds them together, by the utilization of the energy by ATP. Sometimes, the aminoacyl -t RNA synthetase makes an error, it can bind to the incorrect amino acid that seems to be like a correct target. For instance, threonine synthetase occasionally picks serine accidently and links it to the threonine. Threonine synthetase has a site that proofreads, which throws the amino acid away from the t RNA if it is not correct.
Translation takes place in three steps; Initiation, Elongation, Termination [31].
Initiation is the initial stage in the procedure of translation .In this step the initiation complex is formed when the mRNA attaches with the ribosome and an initiator t RNA which carries the initial amino acid making protein, that is usually methionine (Met). Inside the cells of the eukaryotes initiation step of translation goes in the way as described below; firstly the t RNA carrying methionine will bind to the smaller subunit of the ribosome probably 40S in size now this structure (of t RNA and smaller subunit of ribosome) will bind to the 5’ cap of the mRNA this cap was added in the nucleus when mRNA was being processed before going into the cytoplasm. Then they start moving in the 3’ direction on the mRNA and stop on reaching the start codon (usually it is AUG, but it is not necessary that always AUG will be the start codon) [32]. GTP (Guanosine triphosphate) a purine nucleotide triphosphate acts as the source of energy during the phenomenon of the translation .It provides energy for starting of the translation and for the transport of the ribosome on the mRNA strand [33].
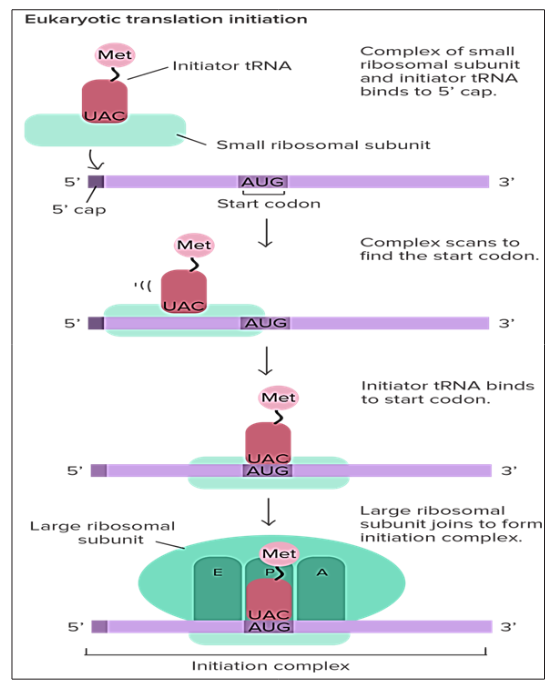
Figure 7: Formation of the initiation complex in Eukaryotes: The T RNA having the anticodon for the first amino acid (Met in case of eukaryotes) binds to the smaller ribosomal subunit, after which it binds to the mRNA, the larger ribosomal subunit is attached to the smaller ribosomal subunit utilizing the energy from GTP.As a result complete ribosome gets attach to the ribosome in two steps .The ribosome moves on the mRNA until or unless it encounters any stop codon /p>
In bacteria, translation takes place in a little different way. In bacteria the ribosome does not attach to the mRNA and move in the 3’ direction rather it directly binds to the certain sequences on the mRNA. These sequences are referred to as the shine-Dalgarno, these are the sequences that are present before the start codons and direct them to the ribosome [34].
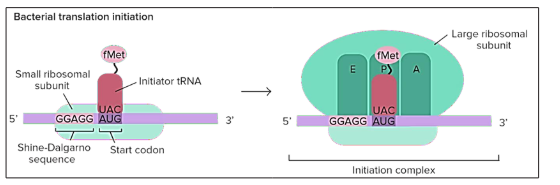
Figure 8: Translational initiation in bacteria: Ribosomes are not moving on the mRNA rather they get attached to the sequence directly and that sequence is Shine -Dalgarno sequence(comprising of 5 nucleotides GGAGG)
Genes of the bacteria are transcribed in the sets called the operons, so single mRNA in the bacteria has the sequences coding for different genes. A shine-Dalgarno sequence is the beginning of the each coding sequence and let the ribosome to mark the correct start codon for each gene.
Elongation is the second step after initiation. It is a very coordinated and multifactorial process [35].The initiation complex is formed by now, methionine which is holding t RNA is present in the central slot on the ribosome (p site), the A site on the ribosome provides the landing site for the another t RNA which carries another amino acids and its anticodon is complementary to the exposed codon. Now a peptide bond is developed between the one amino acid present on the tRNA on p site and another amino acid present on t RNA at A site. So by now a polypeptide comprising of two amino acids is achieved. Making of the peptide bond between the methionine and another amino acid results in the detachment of the methionine from the t RNA present at P site and gets bound to the amino acid present at t RNA on A site. Now as the making of the peptide bond is completed the mRNA is moved forward along the ribosome by precisely one codon. This movement permit the unoccupied t RNA to move out through the E (exit) site. The new codon is also exposed on the A site so that the whole process of the elongation is repeated for the second time to increase the number of amino acid in the polypeptide chain. As a result the polypeptide chain is made in the direction from terminal N to the terminal C (N terminal is the exposed amino group NH2- and the C terminal is the exposed carboxyl group COOH-).This elongation cycle can be repeated several times approximately, 33, ooo times usually. A protein known as titin which is found in muscles of animals is the largest protein. The energy for each step of the ribosome is donated by an elongation factor that hydrolyses the GTP. The making of the peptide bond between amino group of the amino acid present at A site of ribosome and the carboxyl group of the amino acid binds to the p-site, t RNA is catalyzed by the peptidyl transferases.
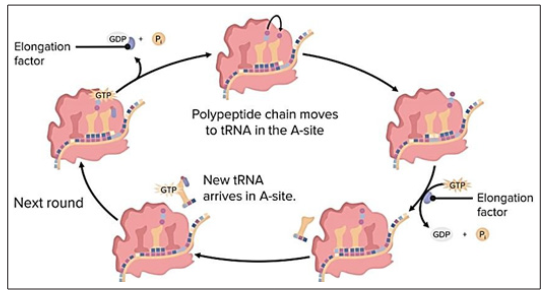
Figure 9: Process of elongation: t RNA on the p site carries the amino acid, until the A site on ribosome attaches another t RNA carryi+ng another amino acid. The amino acid present on the tRNA at p site for making a peptide bond along with the amino acid present on A site, thus the empty t RNA at p site departs through the exit site
The process of translation eventually ends and this step is referred to as termination. When the stop codon which is present in the mRNA take up the A site on ribosome, this step is known as termination. Release factors are the proteins which are used to recognize the stop codons. These proteins fix accurately inside the p site although they are not the t RNA. Release factors disturb the enzymes that form peptide bonds; they hold up a molecule of water to the amino acid present at last in the chain. As a consequence of this phenomenon the t RNA is segregated from the chain and protein which was synthesized newly is released. After the process of the translation ends up the large and the smaller subunits of ribosome separate from the mRNA .The machinery for the translation process is reusable and can take part in another round of translation [36].
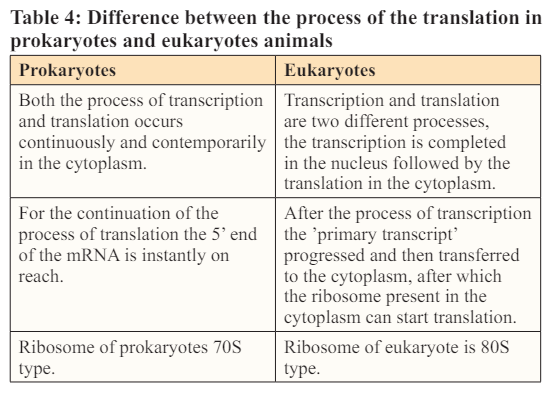
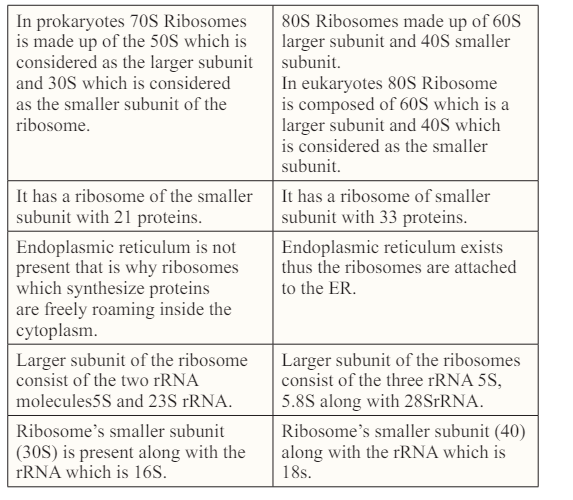
Many changes takes place in the polypeptides after the initiation of their synthesis. Most frequently these modifications include protein folding, trimming by proteolytic degradation, intein splicing and covalent changes which are collectively known as post translational modifications.
The process of expression of gene in prokaryotic and eukaryotic cells is very important as it leads to the formation of the protein, which in turn forms the whole body. Gene expression is a vast topic and day-by-day new discoveries are made in this field to explore the mechanism of formation of protein which is a very important molecule and any mutations in this process can lead to some very adverse results.
Authors express their sincere gratitude for giving support to publish this paper to Prof. G. Paddaiah, Department of Human Genetics, Andhra University, Visakhapatnam, Andhra Pradesh, India and the Authors express their sincere regards for encouraging, to Prof. K. Padma Raju, Vice Chancellor, Adikavi Nannaya University, Rajamahendravaram, East Godavari District, Andhra Pradesh, India.
